bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2442_orf1
Length=97
Score E
Sequences producing significant alignments: (Bits) Value
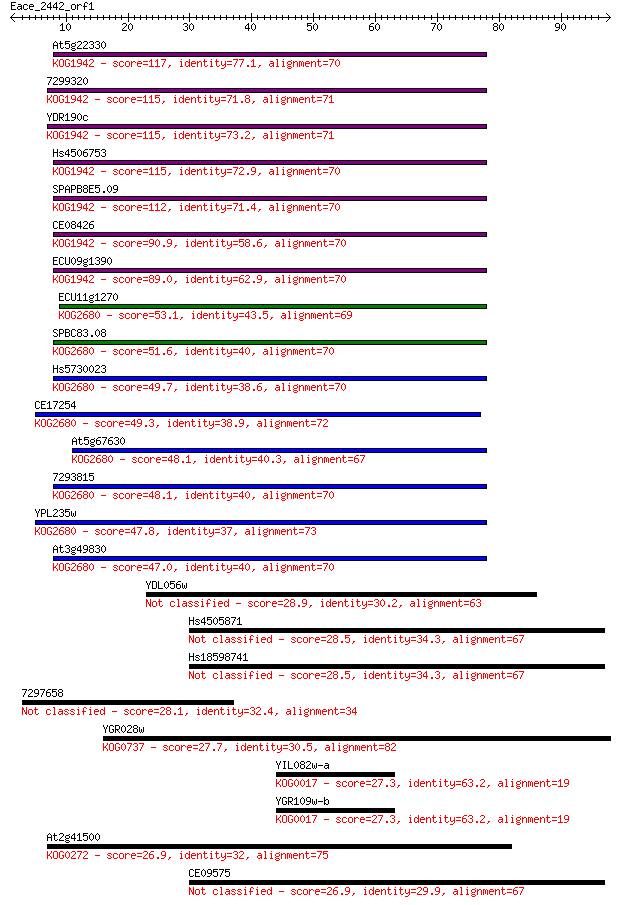
At5g22330 117 7e-27
7299320 115 2e-26
YDR190c 115 2e-26
Hs4506753 115 3e-26
SPAPB8E5.09 112 1e-25
CE08426 90.9 5e-19
ECU09g1390 89.0 2e-18
ECU11g1270 53.1 1e-07
SPBC83.08 51.6 3e-07
Hs5730023 49.7 1e-06
CE17254 49.3 2e-06
At5g67630 48.1 3e-06
7293815 48.1 4e-06
YPL235w 47.8 6e-06
At3g49830 47.0 9e-06
YDL056w 28.9 2.2
Hs4505871 28.5 3.3
Hs18598741 28.5 3.5
7297658 28.1 4.7
YGR028w 27.7 4.8
YIL082w-a 27.3 7.4
YGR109w-b 27.3 7.7
At2g41500 26.9 8.3
CE09575 26.9 9.6
> At5g22330
Length=458
Score = 117 bits (292), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 54/70 (77%), Positives = 62/70 (88%), Gaps = 0/70 (0%)
Query 8 LQDVTLHDLDAANAKPSGGAPLASLLGHFSKPRKTEITEKLRQEINKVVNKYLDQGVAEL 67
+QDVTL DLDAANA+P GG + SL+G KPRKTEIT+KLRQEINKVVN+Y+D+GVAEL
Sbjct 238 VQDVTLQDLDAANARPQGGQDILSLMGQMMKPRKTEITDKLRQEINKVVNRYIDEGVAEL 297
Query 68 VPGVLFIDEV 77
VPGVLFIDEV
Sbjct 298 VPGVLFIDEV 307
> 7299320
Length=456
Score = 115 bits (288), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 51/71 (71%), Positives = 61/71 (85%), Gaps = 0/71 (0%)
Query 7 CLQDVTLHDLDAANAKPSGGAPLASLLGHFSKPRKTEITEKLRQEINKVVNKYLDQGVAE 66
+QDVTLHDLD ANA+P GG + S++G KP+KTEIT+KLR EINKVVNKY+DQG+AE
Sbjct 234 VIQDVTLHDLDVANARPQGGQDVLSMMGQLMKPKKTEITDKLRMEINKVVNKYIDQGIAE 293
Query 67 LVPGVLFIDEV 77
LVPGVLFIDE+
Sbjct 294 LVPGVLFIDEI 304
> YDR190c
Length=463
Score = 115 bits (288), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 52/71 (73%), Positives = 61/71 (85%), Gaps = 0/71 (0%)
Query 7 CLQDVTLHDLDAANAKPSGGAPLASLLGHFSKPRKTEITEKLRQEINKVVNKYLDQGVAE 66
+QDVTLHDLD ANA+P GG + S++G KP+KTEITEKLRQE+NKVV KY+DQGVAE
Sbjct 243 IVQDVTLHDLDVANARPQGGQDVISMMGQLLKPKKTEITEKLRQEVNKVVAKYIDQGVAE 302
Query 67 LVPGVLFIDEV 77
L+PGVLFIDEV
Sbjct 303 LIPGVLFIDEV 313
> Hs4506753
Length=456
Score = 115 bits (287), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 51/70 (72%), Positives = 61/70 (87%), Gaps = 0/70 (0%)
Query 8 LQDVTLHDLDAANAKPSGGAPLASLLGHFSKPRKTEITEKLRQEINKVVNKYLDQGVAEL 67
+QDVTLHDLD ANA+P GG + S++G KP+KTEIT+KLR EINKVVNKY+DQG+AEL
Sbjct 235 IQDVTLHDLDVANARPQGGQDILSMMGQLMKPKKTEITDKLRGEINKVVNKYIDQGIAEL 294
Query 68 VPGVLFIDEV 77
VPGVLF+DEV
Sbjct 295 VPGVLFVDEV 304
> SPAPB8E5.09
Length=456
Score = 112 bits (281), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 50/70 (71%), Positives = 61/70 (87%), Gaps = 0/70 (0%)
Query 8 LQDVTLHDLDAANAKPSGGAPLASLLGHFSKPRKTEITEKLRQEINKVVNKYLDQGVAEL 67
+QDVTLHDLD ANA+P GG + S++G KP+KTEIT+KLR EINKVVNKY++QG+AEL
Sbjct 236 VQDVTLHDLDIANARPQGGQDIMSMMGQLMKPKKTEITDKLRGEINKVVNKYIEQGIAEL 295
Query 68 VPGVLFIDEV 77
+PGVLFIDEV
Sbjct 296 IPGVLFIDEV 305
> CE08426
Length=458
Score = 90.9 bits (224), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 41/71 (57%), Positives = 58/71 (81%), Gaps = 1/71 (1%)
Query 8 LQDVTLHDLDAANAKPSG-GAPLASLLGHFSKPRKTEITEKLRQEINKVVNKYLDQGVAE 66
+Q+V+LHDLD ANA+P G +++++ P+KTE+T++LR EINKVVN+Y++ GVAE
Sbjct 253 VQNVSLHDLDIANARPQGRQGDVSNIVSQLMTPKKTEVTDRLRSEINKVVNEYIESGVAE 312
Query 67 LVPGVLFIDEV 77
L+PGVLFIDEV
Sbjct 313 LMPGVLFIDEV 323
> ECU09g1390
Length=426
Score = 89.0 bits (219), Expect = 2e-18, Method: Composition-based stats.
Identities = 44/70 (62%), Positives = 55/70 (78%), Gaps = 1/70 (1%)
Query 8 LQDVTLHDLDAANAKPSGGAPLASLLGHFSKPRKTEITEKLRQEINKVVNKYLDQGVAEL 67
+Q VTLHDLD ANA+PSG + SL+ PRKTEITE+LR ++N++VN YL+ G AE+
Sbjct 227 MQSVTLHDLDMANARPSG-QDMLSLVFRILSPRKTEITERLRGDVNRMVNGYLENGNAEI 285
Query 68 VPGVLFIDEV 77
VPGVLFIDEV
Sbjct 286 VPGVLFIDEV 295
> ECU11g1270
Length=418
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 30/69 (43%), Positives = 42/69 (60%), Gaps = 7/69 (10%)
Query 9 QDVTLHDLDAANAKPSGGAPLASLLGHFSKPRKTEITEKLRQEINKVVNKYLDQGVAELV 68
Q+++LHD+D N+K G L FS EI + R E+NK V ++++G AE+V
Sbjct 214 QEISLHDIDVVNSKAEG------YLALFSG-ETGEIRAETRDEVNKKVWGWINEGKAEIV 266
Query 69 PGVLFIDEV 77
GVLFIDEV
Sbjct 267 RGVLFIDEV 275
> SPBC83.08
Length=465
Score = 51.6 bits (122), Expect = 3e-07, Method: Composition-based stats.
Identities = 28/70 (40%), Positives = 44/70 (62%), Gaps = 7/70 (10%)
Query 8 LQDVTLHDLDAANAKPSGGAPLASLLGHFSKPRKTEITEKLRQEINKVVNKYLDQGVAEL 67
+ V+LHD+D N++ G L FS EI ++R++IN V+++ ++G AE+
Sbjct 233 VHTVSLHDIDVINSRTQG------FLALFSGD-TGEIKPEVREQINTKVSEWREEGKAEI 285
Query 68 VPGVLFIDEV 77
VPGVLF+DEV
Sbjct 286 VPGVLFVDEV 295
> Hs5730023
Length=463
Score = 49.7 bits (117), Expect = 1e-06, Method: Composition-based stats.
Identities = 27/70 (38%), Positives = 43/70 (61%), Gaps = 7/70 (10%)
Query 8 LQDVTLHDLDAANAKPSGGAPLASLLGHFSKPRKTEITEKLRQEINKVVNKYLDQGVAEL 67
+ V+LH++D N++ G L FS EI ++R++IN V ++ ++G AE+
Sbjct 239 VHTVSLHEIDVINSRTQG------FLALFSGD-TGEIKSEVREQINAKVAEWREEGKAEI 291
Query 68 VPGVLFIDEV 77
+PGVLFIDEV
Sbjct 292 IPGVLFIDEV 301
> CE17254
Length=448
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 28/72 (38%), Positives = 40/72 (55%), Gaps = 7/72 (9%)
Query 5 RLCLQDVTLHDLDAANAKPSGGAPLASLLGHFSKPRKTEITEKLRQEINKVVNKYLDQGV 64
R + V LHD+D N++ G L S EI ++R +INK V ++ ++G
Sbjct 232 RETVHTVCLHDIDVINSRTQGYVALFS-------GDTGEIKAEVRDQINKKVLEWREEGK 284
Query 65 AELVPGVLFIDE 76
A+ VPGVLFIDE
Sbjct 285 AKFVPGVLFIDE 296
> At5g67630
Length=469
Score = 48.1 bits (113), Expect = 3e-06, Method: Composition-based stats.
Identities = 27/67 (40%), Positives = 42/67 (62%), Gaps = 7/67 (10%)
Query 11 VTLHDLDAANAKPSGGAPLASLLGHFSKPRKTEITEKLRQEINKVVNKYLDQGVAELVPG 70
VTLH++D N++ G L F+ EI ++R++I+ V ++ ++G AE+VPG
Sbjct 239 VTLHEIDVINSRTQG------FLALFTGD-TGEIRSEVREQIDTKVAEWREEGKAEIVPG 291
Query 71 VLFIDEV 77
VLFIDEV
Sbjct 292 VLFIDEV 298
> 7293815
Length=481
Score = 48.1 bits (113), Expect = 4e-06, Method: Composition-based stats.
Identities = 28/70 (40%), Positives = 42/70 (60%), Gaps = 7/70 (10%)
Query 8 LQDVTLHDLDAANAKPSGGAPLASLLGHFSKPRKTEITEKLRQEINKVVNKYLDQGVAEL 67
+ VTLH++D N++ G L FS EI +++R +IN V ++ ++G AE+
Sbjct 235 VHTVTLHEIDVINSRTHG------FLALFSGD-TGEIKQEVRDQINNKVLEWREEGKAEI 287
Query 68 VPGVLFIDEV 77
PGVLFIDEV
Sbjct 288 NPGVLFIDEV 297
> YPL235w
Length=471
Score = 47.8 bits (112), Expect = 6e-06, Method: Composition-based stats.
Identities = 27/73 (36%), Positives = 43/73 (58%), Gaps = 7/73 (9%)
Query 5 RLCLQDVTLHDLDAANAKPSGGAPLASLLGHFSKPRKTEITEKLRQEINKVVNKYLDQGV 64
+ + V+LH++D N++ G L F+ EI ++R +IN V ++ ++G
Sbjct 233 KTVVHTVSLHEIDVINSRTQG------FLALFTGD-TGEIRSEVRDQINTKVAEWKEEGK 285
Query 65 AELVPGVLFIDEV 77
AE+VPGVLFIDEV
Sbjct 286 AEIVPGVLFIDEV 298
> At3g49830
Length=473
Score = 47.0 bits (110), Expect = 9e-06, Method: Composition-based stats.
Identities = 28/70 (40%), Positives = 41/70 (58%), Gaps = 7/70 (10%)
Query 8 LQDVTLHDLDAANAKPSGGAPLASLLGHFSKPRKTEITEKLRQEINKVVNKYLDQGVAEL 67
L VTLH++D N++ G LA G EI + R++ + V ++ ++G AE+
Sbjct 237 LHSVTLHEIDVINSRTQG--YLALFTGD-----TGEIRSETREQSDTKVAEWREEGKAEI 289
Query 68 VPGVLFIDEV 77
VPGVLFIDEV
Sbjct 290 VPGVLFIDEV 299
> YDL056w
Length=833
Score = 28.9 bits (63), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 38/67 (56%), Gaps = 8/67 (11%)
Query 23 PSGG-APLASLLGHF---SKPRKTEITEKLRQEINKVVNKYLDQGVAELVPGVLFIDEVL 78
P GG +P+ S++ + S+P+ ++I +K +NK ++K +D ++ + + +VL
Sbjct 321 PGGGTSPIISMIPRYPVTSRPQTSDINDK----VNKYLSKLVDYFISNEMKSNKSLPQVL 376
Query 79 QHPRPSS 85
HP P S
Sbjct 377 LHPPPHS 383
> Hs4505871
Length=1252
Score = 28.5 bits (62), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 34/73 (46%), Gaps = 6/73 (8%)
Query 30 ASLLGHFSKPRKTEITEKLRQEINKV---VNKYLDQGVAELVPGVLFIDEVLQH--PRPS 84
A L F + E E Q++NKV + K++D + E LF+DE L + R +
Sbjct 240 AVYLHDFQRFLIHEQQEHWAQDLNKVRERMTKFIDDTMRETAEPFLFVDEFLTYLFSREN 299
Query 85 S-TSPSYDRVTLQ 96
S YD V +Q
Sbjct 300 SIWDEKYDAVDMQ 312
> Hs18598741
Length=1265
Score = 28.5 bits (62), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 34/73 (46%), Gaps = 6/73 (8%)
Query 30 ASLLGHFSKPRKTEITEKLRQEINKV---VNKYLDQGVAELVPGVLFIDEVLQH--PRPS 84
A L F + E E Q++NKV + K++D + E LF+DE L + R +
Sbjct 240 AVYLHDFQRFLIHEQQEHWAQDLNKVRERMTKFIDDTMRETAEPFLFVDEFLTYLFSREN 299
Query 85 S-TSPSYDRVTLQ 96
S YD V +Q
Sbjct 300 SIWDEKYDAVDMQ 312
> 7297658
Length=627
Score = 28.1 bits (61), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 11/34 (32%), Positives = 18/34 (52%), Gaps = 0/34 (0%)
Query 3 HIRLCLQDVTLHDLDAANAKPSGGAPLASLLGHF 36
HI +C + +D++ N KP+G P+A H
Sbjct 291 HITVCSEQKPNNDVNQVNKKPNGTIPVAITEEHL 324
> YGR028w
Length=362
Score = 27.7 bits (60), Expect = 4.8, Method: Composition-based stats.
Identities = 25/82 (30%), Positives = 37/82 (45%), Gaps = 9/82 (10%)
Query 16 LDAANAKPSGGAPLASLLGHFSKPRKTEITEKLRQEINKVVNKYLDQGVAELVPGVLFID 75
L A AK SG +F R + I +K E NK+V+ +L P ++FID
Sbjct 142 LAKALAKESGA--------NFISIRMSSIMDKWYGESNKIVDAMFSLA-NKLQPCIIFID 192
Query 76 EVLQHPRPSSTSPSYDRVTLQA 97
E+ R S++ TL+A
Sbjct 193 EIDSFLRERSSTDHEVTATLKA 214
> YIL082w-a
Length=1498
Score = 27.3 bits (59), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 12/19 (63%), Positives = 14/19 (73%), Gaps = 0/19 (0%)
Query 44 ITEKLRQEINKVVNKYLDQ 62
+TEK QEINK+V K LD
Sbjct 630 VTEKNEQEINKIVQKLLDN 648
> YGR109w-b
Length=1547
Score = 27.3 bits (59), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 12/19 (63%), Positives = 14/19 (73%), Gaps = 0/19 (0%)
Query 44 ITEKLRQEINKVVNKYLDQ 62
+TEK QEINK+V K LD
Sbjct 604 VTEKNEQEINKIVQKLLDN 622
> At2g41500
Length=554
Score = 26.9 bits (58), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 24/87 (27%), Positives = 40/87 (45%), Gaps = 13/87 (14%)
Query 7 CLQDVTLHDLDAANAKPSG-----------GAPLASLLGHFSKPRKTEITEKL-RQEINK 54
LQD+ + AA A P+ G P+ +L G R+ +T+ L R +IN
Sbjct 104 ALQDLLVKRRAAAMAVPTNDKAVRDRLRRLGEPI-TLFGEQEMERRARLTQLLTRYDING 162
Query 55 VVNKYLDQGVAELVPGVLFIDEVLQHP 81
++K + ++ P DEVL++P
Sbjct 163 QLDKLVKDHEEDVTPKEEVDDEVLEYP 189
> CE09575
Length=219
Score = 26.9 bits (58), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 20/77 (25%), Positives = 36/77 (46%), Gaps = 10/77 (12%)
Query 30 ASLLGHFSKP--RKTEITEKLRQEINKVVNK---YLD-----QGVAELVPGVLFIDEVLQ 79
+S+ GHFS R ++E+ Q + + + Y + G A+ VP +E+
Sbjct 34 SSICGHFSSTIRRYHHVSEREAQRFQQQLRQPGCYQECLKRKTGTAKEVPAKKAKEEIFA 93
Query 80 HPRPSSTSPSYDRVTLQ 96
P PS +PS D + ++
Sbjct 94 KPSPSKPAPSADTMGIE 110
Lambda K H
0.318 0.134 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1198045380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40