bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2343_orf1
Length=155
Score E
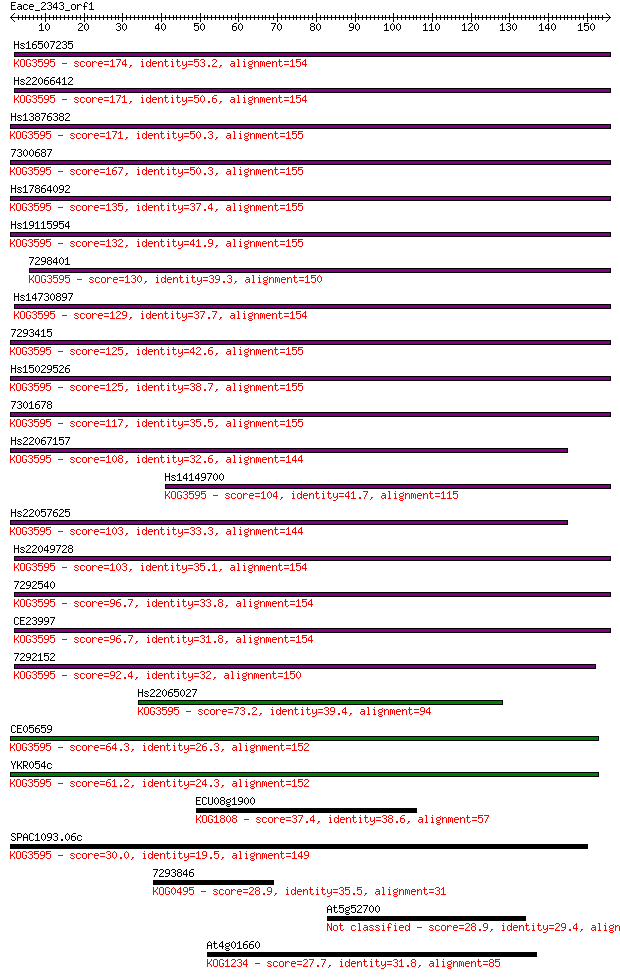
Sequences producing significant alignments: (Bits) Value
Hs16507235 174 8e-44
Hs22066412 171 4e-43
Hs13876382 171 7e-43
7300687 167 8e-42
Hs17864092 135 4e-32
Hs19115954 132 3e-31
7298401 130 1e-30
Hs14730897 129 3e-30
7293415 125 2e-29
Hs15029526 125 4e-29
7301678 117 1e-26
Hs22067157 108 4e-24
Hs14149700 104 6e-23
Hs22057625 103 2e-22
Hs22049728 103 2e-22
7292540 96.7 2e-20
CE23997 96.7 2e-20
7292152 92.4 3e-19
Hs22065027 73.2 2e-13
CE05659 64.3 9e-11
YKR054c 61.2 8e-10
ECU08g1900 37.4 0.012
SPAC1093.06c 30.0 1.8
7293846 28.9 3.7
At5g52700 28.9 4.7
At4g01660 27.7 9.4
> Hs16507235
Length=4523
Score = 174 bits (440), Expect = 8e-44, Method: Compositional matrix adjust.
Identities = 82/154 (53%), Positives = 111/154 (72%), Gaps = 2/154 (1%)
Query 2 ENAAIVNSCMRWPLLIDPQLQGTRWVKQRGRDSLITVSVNRDRWLNKVIDAIRSGDVLLI 61
ENAAI+ C RWPL+IDPQ QG +W+K + L + + +LN + A+ GDV+LI
Sbjct 3488 ENAAILTHCERWPLVIDPQQQGIKWIKNKYGMDLKVTHLGQKGFLNAIETALAFGDVILI 3547
Query 62 ENLSEAIDPVLDPLVSRSVSRKGRTLYVKMGGEDVEFSSSFRLLLQTKLPNPHYKPEIAA 121
ENL E IDPVLDPL+ R+ +KG+ Y+++G ++ EF+ +FRL+L TKL NPHYKPE+ A
Sbjct 3548 ENLEETIDPVLDPLLGRNTIKKGK--YIRIGDKECEFNKNFRLILHTKLANPHYKPELQA 3605
Query 122 QCTLVNFTVTPEGLEEQFLAMIVNAEQPDLETSK 155
Q TL+NFTVT +GLE Q LA +V+ E+PDLE K
Sbjct 3606 QTTLLNFTVTEDGLEAQLLAEVVSIERPDLEKLK 3639
> Hs22066412
Length=667
Score = 171 bits (434), Expect = 4e-43, Method: Compositional matrix adjust.
Identities = 78/154 (50%), Positives = 109/154 (70%), Gaps = 2/154 (1%)
Query 2 ENAAIVNSCMRWPLLIDPQLQGTRWVKQRGRDSLITVSVNRDRWLNKVIDAIRSGDVLLI 61
ENA I+ + RWPL++D QLQG +W+K + R L + + + +L+ + AI GD LLI
Sbjct 238 ENATILGNTERWPLIVDAQLQGIKWIKNKYRSELKAIRLGQKSYLDVIEQAISEGDTLLI 297
Query 62 ENLSEAIDPVLDPLVSRSVSRKGRTLYVKMGGEDVEFSSSFRLLLQTKLPNPHYKPEIAA 121
EN+ E +DPVLDPL+ R+ +KG+ Y+K+G ++VE+ FRL+L TK NPHYKPE+ A
Sbjct 298 ENIGETVDPVLDPLLGRNTIKKGK--YIKIGDKEVEYHPKFRLILHTKYFNPHYKPEMQA 355
Query 122 QCTLVNFTVTPEGLEEQFLAMIVNAEQPDLETSK 155
QCTL+NF VT +GLE+Q LA +V E+PDLE K
Sbjct 356 QCTLINFLVTRDGLEDQLLAAVVAKERPDLEQLK 389
> Hs13876382
Length=4486
Score = 171 bits (432), Expect = 7e-43, Method: Compositional matrix adjust.
Identities = 78/155 (50%), Positives = 113/155 (72%), Gaps = 2/155 (1%)
Query 1 IENAAIVNSCMRWPLLIDPQLQGTRWVKQRGRDSLITVSVNRDRWLNKVIDAIRSGDVLL 60
+ENA I+ +C RWPL++DPQLQG +W+K + + L + + +L + A+ +G V+L
Sbjct 3450 VENATILINCERWPLMVDPQLQGIKWIKNKYGEDLRVTQIGQKGYLQIIEQALEAGAVVL 3509
Query 61 IENLSEAIDPVLDPLVSRSVSRKGRTLYVKMGGEDVEFSSSFRLLLQTKLPNPHYKPEIA 120
IENL E+IDPVL PL+ R V +KGR ++K+G ++ E++ FRL+L TKL NPHY+PE+
Sbjct 3510 IENLEESIDPVLGPLLGREVIKKGR--FIKIGDKECEYNPKFRLILHTKLANPHYQPELQ 3567
Query 121 AQCTLVNFTVTPEGLEEQFLAMIVNAEQPDLETSK 155
AQ TL+NFTVT +GLE+Q LA +V+ E+PDLE K
Sbjct 3568 AQATLINFTVTRDGLEDQLLAAVVSMERPDLEQLK 3602
> 7300687
Length=4472
Score = 167 bits (423), Expect = 8e-42, Method: Compositional matrix adjust.
Identities = 78/155 (50%), Positives = 117/155 (75%), Gaps = 2/155 (1%)
Query 1 IENAAIVNSCMRWPLLIDPQLQGTRWVKQRGRDSLITVSVNRDRWLNKVIDAIRSGDVLL 60
IENA I+++ RWPL+IDPQLQG +W+KQ+ + L + + + +L+ + +I +G +L
Sbjct 3435 IENATILSNSDRWPLMIDPQLQGVKWIKQKYGEDLKVIRLGQRSYLDIIEKSINAGCNVL 3494
Query 61 IENLSEAIDPVLDPLVSRSVSRKGRTLYVKMGGEDVEFSSSFRLLLQTKLPNPHYKPEIA 120
IEN+ E +DPVLD L+ R++ +KG+ +K+G +++E++S+FRL+L TKL NPHYKPE+
Sbjct 3495 IENIDENLDPVLDSLLGRNLIKKGKA--IKIGDKEIEYNSNFRLILHTKLANPHYKPEMQ 3552
Query 121 AQCTLVNFTVTPEGLEEQFLAMIVNAEQPDLETSK 155
AQ TL+NFTVT +GLE+Q LA +V AE+PDLE K
Sbjct 3553 AQTTLINFTVTRDGLEDQLLAEVVKAERPDLEELK 3587
> Hs17864092
Length=4024
Score = 135 bits (339), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 58/156 (37%), Positives = 102/156 (65%), Gaps = 1/156 (0%)
Query 1 IENAAIVNSCMRWPLLIDPQLQGTRWVKQRGR-DSLITVSVNRDRWLNKVIDAIRSGDVL 59
I+N I+ + RWPL+IDPQ Q +W+K + +SL + ++ ++ + + I+ G +
Sbjct 2975 IDNGIIIMNARRWPLMIDPQSQANKWIKNMEKANSLYVIKLSEPDYVRTLENCIQFGTPV 3034
Query 60 LIENLSEAIDPVLDPLVSRSVSRKGRTLYVKMGGEDVEFSSSFRLLLQTKLPNPHYKPEI 119
L+EN+ E +DP+L+PL+ + ++G + +++G +E++ FR + TKL NPHY PE
Sbjct 3035 LLENVGEELDPILEPLLLKQTFKQGGSTCIRLGDSTIEYAPDFRFYITTKLRNPHYLPET 3094
Query 120 AAQCTLVNFTVTPEGLEEQFLAMIVNAEQPDLETSK 155
+ + TL+NF +TPEG+++Q L ++V E+PDLE K
Sbjct 3095 SVKVTLLNFMITPEGMQDQLLGIVVAQERPDLEEEK 3130
> Hs19115954
Length=4624
Score = 132 bits (332), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 65/156 (41%), Positives = 100/156 (64%), Gaps = 1/156 (0%)
Query 1 IENAAIVNSCMRWPLLIDPQLQGTRWVKQR-GRDSLITVSVNRDRWLNKVIDAIRSGDVL 59
I+N IV R+PLLIDPQ QG W+K + R+ L S+N + N + D++ G L
Sbjct 3588 IQNGIIVTKASRYPLLIDPQTQGKIWIKNKESRNELQITSLNHKYFRNHLEDSLSLGRPL 3647
Query 60 LIENLSEAIDPVLDPLVSRSVSRKGRTLYVKMGGEDVEFSSSFRLLLQTKLPNPHYKPEI 119
LIE++ E +DP LD ++ R+ + G T VK+G ++V+ FRL + TKLPNP Y PEI
Sbjct 3648 LIEDVGEELDPALDNVLERNFIKTGSTFKVKVGDKEVDVLDGFRLYITTKLPNPAYTPEI 3707
Query 120 AAQCTLVNFTVTPEGLEEQFLAMIVNAEQPDLETSK 155
+A+ ++++FTVT +GLE+Q L ++ E+ +LE +
Sbjct 3708 SARTSIIDFTVTMKGLEDQLLGRVILTEKQELEKER 3743
> 7298401
Length=4010
Score = 130 bits (326), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 59/151 (39%), Positives = 100/151 (66%), Gaps = 1/151 (0%)
Query 6 IVNSCMRWPLLIDPQLQGTRWVKQRGRDS-LITVSVNRDRWLNKVIDAIRSGDVLLIENL 64
I + RWPL+IDPQ Q +W+K +++ L + +N+ + + +AI+ G +L+EN+
Sbjct 2953 ICRNARRWPLMIDPQGQANKWIKNYEKNNKLCVIRLNQADYTRVMENAIQFGLPVLLENI 3012
Query 65 SEAIDPVLDPLVSRSVSRKGRTLYVKMGGEDVEFSSSFRLLLQTKLPNPHYKPEIAAQCT 124
E +DPVL+ ++ +++ ++G L +K+G +E++ SFR + TKL NPHY PE+A + T
Sbjct 3013 GEELDPVLESVLQKTLFKQGGALCIKLGDSVIEYNHSFRFYMTTKLRNPHYLPEVAVKVT 3072
Query 125 LVNFTVTPEGLEEQFLAMIVNAEQPDLETSK 155
L+NF +T +GL++Q L + V E+PDLE K
Sbjct 3073 LLNFMITTQGLQDQLLGITVARERPDLEAEK 3103
> Hs14730897
Length=1350
Score = 129 bits (324), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 58/155 (37%), Positives = 96/155 (61%), Gaps = 1/155 (0%)
Query 2 ENAAIVNSCMRWPLLIDPQLQGTRWVKQR-GRDSLITVSVNRDRWLNKVIDAIRSGDVLL 60
EN +V RWPL+IDPQ Q RW++ + + L + + +L + ++IR G +L
Sbjct 256 ENGILVTQGRRWPLMIDPQDQANRWIRNKESKSGLKIIKLTDSNFLRILENSIRLGLPVL 315
Query 61 IENLSEAIDPVLDPLVSRSVSRKGRTLYVKMGGEDVEFSSSFRLLLQTKLPNPHYKPEIA 120
+E L E +DP L+P++ + + G L +++G D+++ +FR + TK+PNPHY PE+
Sbjct 316 LEELKETLDPALEPILLKQIFISGGRLLIRLGDSDIDYDKNFRFYMTTKMPNPHYLPEVC 375
Query 121 AQCTLVNFTVTPEGLEEQFLAMIVNAEQPDLETSK 155
+ T++NFTVT GLE+Q L+ +V E+P LE +
Sbjct 376 IKVTIINFTVTKSGLEDQLLSDVVRLEKPRLEEQR 410
> 7293415
Length=4081
Score = 125 bits (315), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 66/157 (42%), Positives = 96/157 (61%), Gaps = 3/157 (1%)
Query 1 IENAAIVNSCMRWPLLIDPQLQGTRWVKQRGRDSLITVSVNRDRWLNKVID-AIRSGDVL 59
IEN +RW L+IDPQ Q RW++ R + + V D + +V++ A+R G +
Sbjct 2998 IENGIYATRALRWALMIDPQEQANRWIRNMERANNLQVIKMTDSTMMRVLENAVRQGYPV 3057
Query 60 LIENLSEAIDPVLDPLVSRSVSR-KGRTLYVKMGGEDVEFSSSFRLLLQTKLPNPHYKPE 118
L+E ++E IDP L P++ R R +GRT Y+K+G +++ +F+L + TKLPNPHY PE
Sbjct 3058 LLEEINETIDPSLRPILQRETYRFEGRT-YLKLGDMVIDYDDNFKLYMTTKLPNPHYLPE 3116
Query 119 IAAQCTLVNFTVTPEGLEEQFLAMIVNAEQPDLETSK 155
+ TLVNF VT GLE+Q LA IV E P +E +
Sbjct 3117 VCINVTLVNFLVTESGLEDQLLADIVAIELPAMEIQR 3153
> Hs15029526
Length=4490
Score = 125 bits (313), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 60/156 (38%), Positives = 100/156 (64%), Gaps = 1/156 (0%)
Query 1 IENAAIVNSCMRWPLLIDPQLQGTRWVKQRGRDSLITV-SVNRDRWLNKVIDAIRSGDVL 59
I+N IV R+PLLIDPQ QG W+K + +++ + V S+NR + + D++ G L
Sbjct 3453 IQNGIIVTKATRYPLLIDPQTQGKTWIKSKEKENDLQVTSLNRKYFRTHLEDSLSLGRPL 3512
Query 60 LIENLSEAIDPVLDPLVSRSVSRKGRTLYVKMGGEDVEFSSSFRLLLQTKLPNPHYKPEI 119
LIE++ E +DP LD ++ ++ + G T VK+G ++ + +F+L + TKLPNP + PEI
Sbjct 3513 LIEDIHEELDPALDNVLEKNFIKSGTTFKVKVGDKECDIMDTFKLYITTKLPNPAFTPEI 3572
Query 120 AAQCTLVNFTVTPEGLEEQFLAMIVNAEQPDLETSK 155
A+ ++++FTVT +GLE Q L ++ E+ +LE +
Sbjct 3573 NAKTSVIDFTVTMKGLENQLLRRVILTEKQELEAER 3608
> 7301678
Length=4820
Score = 117 bits (292), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 55/156 (35%), Positives = 100/156 (64%), Gaps = 1/156 (0%)
Query 1 IENAAIVNSCMRWPLLIDPQLQGTRWVKQRG-RDSLITVSVNRDRWLNKVIDAIRSGDVL 59
I+N + R+PL IDPQLQ +W+++R +++L +S + +L ++ AI G +
Sbjct 3982 IQNGILTMRASRFPLCIDPQLQALQWIRKREFKNNLKVLSFSDSDFLKQLEMAIMYGTPV 4041
Query 60 LIENLSEAIDPVLDPLVSRSVSRKGRTLYVKMGGEDVEFSSSFRLLLQTKLPNPHYKPEI 119
L E++ + IDPV+D ++ +++ +G +V +G ++V++ SFR+ L TK NP + P +
Sbjct 4042 LFEDVDDYIDPVIDDILQKNIRIQGGRKFVMLGDKEVDWDPSFRVYLTTKFSNPKFDPAV 4101
Query 120 AAQCTLVNFTVTPEGLEEQFLAMIVNAEQPDLETSK 155
A+ ++N+TVT GLE+Q L+++V E+PDLE +
Sbjct 4102 YAKALVINYTVTQTGLEDQLLSVVVGTERPDLEAQR 4137
> Hs22067157
Length=3672
Score = 108 bits (270), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 47/145 (32%), Positives = 90/145 (62%), Gaps = 1/145 (0%)
Query 1 IENAAIVNSCMRWPLLIDPQLQGTRWVKQRGR-DSLITVSVNRDRWLNKVIDAIRSGDVL 59
I+N IV++ RW L+IDP Q +W+K + + L + + ++ + +A++ G +
Sbjct 2622 IDNGIIVSNSRRWALMIDPHGQANKWIKNMEKANKLAVIKFSDSNYMRMLENALQLGTPV 2681
Query 60 LIENLSEAIDPVLDPLVSRSVSRKGRTLYVKMGGEDVEFSSSFRLLLQTKLPNPHYKPEI 119
LIEN+ E +D ++P++ ++ ++ Y+++G +E+S F+L + T+L NPHY PE+
Sbjct 2682 LIENIGEELDASIEPILLKATFKQQGVEYMRLGENIIEYSRDFKLYITTRLRNPHYLPEV 2741
Query 120 AAQCTLVNFTVTPEGLEEQFLAMIV 144
A + L+NF +TP GL++Q L ++
Sbjct 2742 AVKVCLLNFMITPLGLQDQLLGIVA 2766
> Hs14149700
Length=892
Score = 104 bits (260), Expect = 6e-23, Method: Compositional matrix adjust.
Identities = 48/115 (41%), Positives = 77/115 (66%), Gaps = 1/115 (0%)
Query 41 NRDRWLNKVIDAIRSGDVLLIENLSEAIDPVLDPLVSRSVSRKGRTLYVKMGGEDVEFSS 100
+RD +L + +AIR G L+EN+ E +DP L+P++ + ++ + +K+G + +
Sbjct 14 DRD-FLRSMENAIRFGKPCLLENVGEELDPALEPVLLKQTYKQQGSTVLKLGDTVIPYHE 72
Query 101 SFRLLLQTKLPNPHYKPEIAAQCTLVNFTVTPEGLEEQFLAMIVNAEQPDLETSK 155
FR+ + TKLPNPHY PEI+ + TL+NFT++P GLE+Q L +V E+PDLE +K
Sbjct 73 DFRMYITTKLPNPHYTPEISTKLTLINFTLSPSGLEDQLLGQVVAEERPDLEEAK 127
> Hs22057625
Length=485
Score = 103 bits (257), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 48/145 (33%), Positives = 89/145 (61%), Gaps = 1/145 (0%)
Query 1 IENAAIVNSCMRWPLLIDPQLQGTRWVKQRG-RDSLITVSVNRDRWLNKVIDAIRSGDVL 59
++N + R+PL IDPQ Q W+K++ +++L S N +L ++ +I+ G
Sbjct 170 VQNGILTTRASRFPLCIDPQQQALNWIKRKEEKNNLRVASFNDPDFLKQLEMSIKYGTPF 229
Query 60 LIENLSEAIDPVLDPLVSRSVSRKGRTLYVKMGGEDVEFSSSFRLLLQTKLPNPHYKPEI 119
L ++ E IDPV+D ++ +++ ++ +G ++V++ S+FRL L TKL NP Y P +
Sbjct 230 LFRDVDEYIDPVIDNVLEKNIKVSQGRQFIILGDKEVDYDSNFRLYLNTKLANPRYSPSV 289
Query 120 AAQCTLVNFTVTPEGLEEQFLAMIV 144
+ ++N+TVT +GLE+Q L+++V
Sbjct 290 FGKAMVINYTVTLKGLEDQLLSVLV 314
> Hs22049728
Length=2274
Score = 103 bits (256), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 54/155 (34%), Positives = 95/155 (61%), Gaps = 2/155 (1%)
Query 2 ENAAIVNSCMRWPLLIDPQLQGTRWVKQRGRDSLITVSVNRDRWLNKVID-AIRSGDVLL 60
ENA ++ R+PL+IDP Q T ++ +D IT + D K ++ A+R G+ LL
Sbjct 1203 ENAIMLKRFNRYPLIIDPSGQATEFIMNEYKDRKITRTSFLDDAFRKNLESALRFGNPLL 1262
Query 61 IENLSEAIDPVLDPLVSRSVSRKGRTLYVKMGGEDVEFSSSFRLLLQTKLPNPHYKPEIA 120
++++ E+ DPVL+P+++R V R G + + +G +D++ S SF + L T+ P + P++
Sbjct 1263 VQDV-ESYDPVLNPVLNREVRRTGGRVLITLGDQDIDLSPSFVIFLSTRDPTVEFPPDLC 1321
Query 121 AQCTLVNFTVTPEGLEEQFLAMIVNAEQPDLETSK 155
++ T VNFTVT L+ Q L ++ AE+PD++ +
Sbjct 1322 SRVTFVNFTVTRSSLQSQCLNEVLKAERPDVDEKR 1356
> 7292540
Length=4680
Score = 96.7 bits (239), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 52/155 (33%), Positives = 91/155 (58%), Gaps = 2/155 (1%)
Query 2 ENAAIVNSCMRWPLLIDPQLQGTRWVKQRGRDSLIT-VSVNRDRWLNKVIDAIRSGDVLL 60
ENA ++ R+PL+IDP Q T ++ IT S D + + A+R G+ LL
Sbjct 3545 ENAIMLKRFNRYPLIIDPSGQATTFLLNEYAGKKITKTSFLDDSFRKNLESALRFGNPLL 3604
Query 61 IENLSEAIDPVLDPLVSRSVSRKGRTLYVKMGGEDVEFSSSFRLLLQTKLPNPHYKPEIA 120
++++ E DP+L+P+++R + R G + + +G +D++ S SF + L T+ P + P+I
Sbjct 3605 VQDV-ENYDPILNPVLNRELRRTGGRVLITLGDQDIDLSPSFVIFLSTRDPTVEFPPDIC 3663
Query 121 AQCTLVNFTVTPEGLEEQFLAMIVNAEQPDLETSK 155
++ T VNFTVT L+ Q L ++ AE+PD++ +
Sbjct 3664 SRVTFVNFTVTRSSLQSQCLNQVLKAERPDIDEKR 3698
> CE23997
Length=4568
Score = 96.7 bits (239), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 49/155 (31%), Positives = 93/155 (60%), Gaps = 2/155 (1%)
Query 2 ENAAIVNSCMRWPLLIDPQLQGTRWV-KQRGRDSLITVSVNRDRWLNKVIDAIRSGDVLL 60
ENA +++ R+PL+IDP Q ++ KQ ++ S + + + A+R G+ LL
Sbjct 3518 ENAIMLHRFNRYPLIIDPSGQAVEYIMKQFAGKNIQKTSFLDESFRKNLESALRFGNSLL 3577
Query 61 IENLSEAIDPVLDPLVSRSVSRKGRTLYVKMGGEDVEFSSSFRLLLQTKLPNPHYKPEIA 120
++++ EA DP+L+P+++R V R G + + +G +D++ S SF++ + T+ + P+I
Sbjct 3578 VQDV-EAYDPILNPVLNREVKRAGGRVLITIGDQDIDLSPSFQIFMITRDSTVEFSPDIC 3636
Query 121 AQCTLVNFTVTPEGLEEQFLAMIVNAEQPDLETSK 155
++ T VNFTVT L Q L ++ +E+PD++ +
Sbjct 3637 SRVTFVNFTVTSSSLASQCLNQVLRSERPDVDKKR 3671
> 7292152
Length=3868
Score = 92.4 bits (228), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 48/151 (31%), Positives = 83/151 (54%), Gaps = 1/151 (0%)
Query 2 ENAAIVNSCMRWPLLIDPQLQGTRWVKQRGR-DSLITVSVNRDRWLNKVIDAIRSGDVLL 60
ENA I + R+ L IDPQ Q W+K R + L V N+ ++ + +A+ G ++
Sbjct 2914 ENAIISANSSRYSLFIDPQAQANNWLKNMERKNRLNCVKFNQSNYMKVIAEALEYGTPVI 2973
Query 61 IENLSEAIDPVLDPLVSRSVSRKGRTLYVKMGGEDVEFSSSFRLLLQTKLPNPHYKPEIA 120
IEN+ E ++ LDP++ R +G ++ +G V + +FRL + L NPH+ PE
Sbjct 2974 IENVQEELEVPLDPILMRQTFVQGGIKHISLGESVVPVNPNFRLYMTCNLRNPHFLPETF 3033
Query 121 AQCTLVNFTVTPEGLEEQFLAMIVNAEQPDL 151
+ T++NF +T L +Q L++++ A+ L
Sbjct 3034 NKVTVINFALTQNALMDQLLSIVILADSKGL 3064
> Hs22065027
Length=187
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 37/99 (37%), Positives = 61/99 (61%), Gaps = 5/99 (5%)
Query 34 SLITVSVNRD-RWLNKVIDAIRSGDVLLIENLSEAIDPVLDPLVSRSVSRKGRTLYVKMG 92
+IT ++ R R +V AI G+ +LIEN+SE IDP+LDP++ ++ YVK+G
Sbjct 87 GVITRAIVRSPRAYRRVTGAIAFGNPILIENISEHIDPILDPILLHQTFKQSGMEYVKLG 146
Query 93 GEDVEFSSSFRLLLQTKLPNPHYKPEIAAQ----CTLVN 127
V++S F+L + T++ NPHY PE++ + C+ +N
Sbjct 147 ENTVQYSHDFKLYITTRMRNPHYLPEVSVKRLNSCSNLN 185
> CE05659
Length=4131
Score = 64.3 bits (155), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 40/152 (26%), Positives = 78/152 (51%), Gaps = 2/152 (1%)
Query 1 IENAAIVNSCMRWPLLIDPQLQGTRWVKQRGRDSLITVSVNRDRWLNKVIDAIRSGDVLL 60
+EN +I+ + PL+ID Q + ++ + S T + + ++ AIR G ++
Sbjct 3121 LENGSILFTSCHAPLIIDRSGQVSLFLSKFLEKSE-TFKAAQPDLMTQIELAIRFGKTII 3179
Query 61 IENLSEAIDPVLDPLVSRSVSRKGRTLYVKMGGEDVEFSSSFRLLLQTKLPNPHYKPEIA 120
I+++ E D L P++ + +S +G + GG+ ++F+ F++ T+ +P
Sbjct 3180 IDDIVE-FDSALIPILRKDLSSQGPRQVISFGGKSIDFNPDFKIYFCTRDEKVDIRPNSY 3238
Query 121 AQCTLVNFTVTPEGLEEQFLAMIVNAEQPDLE 152
Q +VNFT T L Q L + ++ E+P+LE
Sbjct 3239 VQLNIVNFTTTISALSAQLLDVAIHLEKPELE 3270
> YKR054c
Length=4092
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 37/153 (24%), Positives = 78/153 (50%), Gaps = 2/153 (1%)
Query 1 IENAAIV-NSCMRWPLLIDPQLQGTRWVKQRGRDSLITVSVNRDRWLNKVIDAIRSGDVL 59
+EN +IV NS P L+DP + + + +S + ++ ++ +AIR G V+
Sbjct 3391 LENMSIVMNSQDAVPFLLDPSSHMITVISNYYGNKTVLLSFLEEGFVKRLENAIRFGSVV 3450
Query 60 LIENLSEAIDPVLDPLVSRSVSRKGRTLYVKMGGEDVEFSSSFRLLLQTKLPNPHYKPEI 119
+I++ E DP++ L+SR + G + V++G +V+ S F+L + + P+ +
Sbjct 3451 IIQD-GEFFDPIISRLISREFNHAGNRVTVEIGDHEVDVSGDFKLFIHSCDPSGDIPIFL 3509
Query 120 AAQCTLVNFTVTPEGLEEQFLAMIVNAEQPDLE 152
++ LV+F E +E + + + E +++
Sbjct 3510 RSRVRLVHFVTNKESIETRIFDITLTEENAEMQ 3542
> ECU08g1900
Length=2832
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 22/57 (38%), Positives = 33/57 (57%), Gaps = 2/57 (3%)
Query 49 VIDAIRSGDVLLIENLSEAIDPVLDPLVSRSVSRKGRTLYVKMGGEDVEFSSSFRLL 105
++ A++ GD LI+ ++ A D VL+ L SV RTLY+ GE+V FR+L
Sbjct 1013 LVKAMKCGDAFLIDEINLAEDSVLERL--NSVLESQRTLYITETGEEVVAHDGFRIL 1067
> SPAC1093.06c
Length=1889
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 29/151 (19%), Positives = 71/151 (47%), Gaps = 4/151 (2%)
Query 1 IENAAIVNSCMRWPLLIDPQLQGTRWVKQ--RGRDSLITVSVNRDRWLNKVIDAIRSGDV 58
+EN I+ L+IDP Q + +G+ S + +S + + N++ A+ SG
Sbjct 1181 LENIYIIQENKSPLLIIDPSSQILDILPSLYKGKASDL-ISFSNKSFQNQIKLALLSGSA 1239
Query 59 LLIENLSEAIDPVLDPLVSRSVSRKGRTLYVKMGGEDVEFSSSFRLLLQTKLPNPHYKPE 118
++I++ +E D ++PL+ + + + + ++ +++ + + +
Sbjct 1240 IIIKD-AELWDVSIEPLLKPEFFTGSGEVQTTFAKDTITITLPLNIIFFSEVQSNELENK 1298
Query 119 IAAQCTLVNFTVTPEGLEEQFLAMIVNAEQP 149
+ +VNFT++ LE Q L +++ ++P
Sbjct 1299 ASKFMNVVNFTLSISLLETQMLKSVISVQEP 1329
> 7293846
Length=913
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 11/31 (35%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 38 VSVNRDRWLNKVIDAIRSGDVLLIENLSEAI 68
V +NRD+W + I+A +SG V +++ +A+
Sbjct 471 VEINRDQWFQEAIEAEKSGAVNCCQSIVKAV 501
> At5g52700
Length=370
Score = 28.9 bits (63), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 26/51 (50%), Gaps = 3/51 (5%)
Query 83 KGRTLYVKMGGEDVEFSSSFRLLLQTKLPNPHYKPEIAAQCTLVNFTVTPE 133
KG+ ++MGG+ E S ++ +T P Y P + T+ N+ TP+
Sbjct 36 KGKFQKIQMGGKLYEIDKSVDIINRTGKPGKSYMPGLN---TVYNYVTTPK 83
> At4g01660
Length=623
Score = 27.7 bits (60), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 27/96 (28%), Positives = 43/96 (44%), Gaps = 26/96 (27%)
Query 52 AIRSGDVLLIENLSEAIDPVLDPLVSRSVSRKGRTLYVKMGGE-----------DVEFSS 100
A++ G +L I++ S P+L+ L YV+ G + D E S
Sbjct 239 ALKVGQMLSIQDESLVPAPILNALE-----------YVRQGADVMPRSQLNPVLDAELGS 287
Query 101 SFRLLLQTKLPNPHYKPEIAAQCTLVNFTVTPEGLE 136
++ Q+KL + Y+P AA V+ VT +GLE
Sbjct 288 NW----QSKLTSFDYEPLAAASIGQVHRAVTKDGLE 319
Lambda K H
0.318 0.135 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2033998736
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40