bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2338_orf1
Length=150
Score E
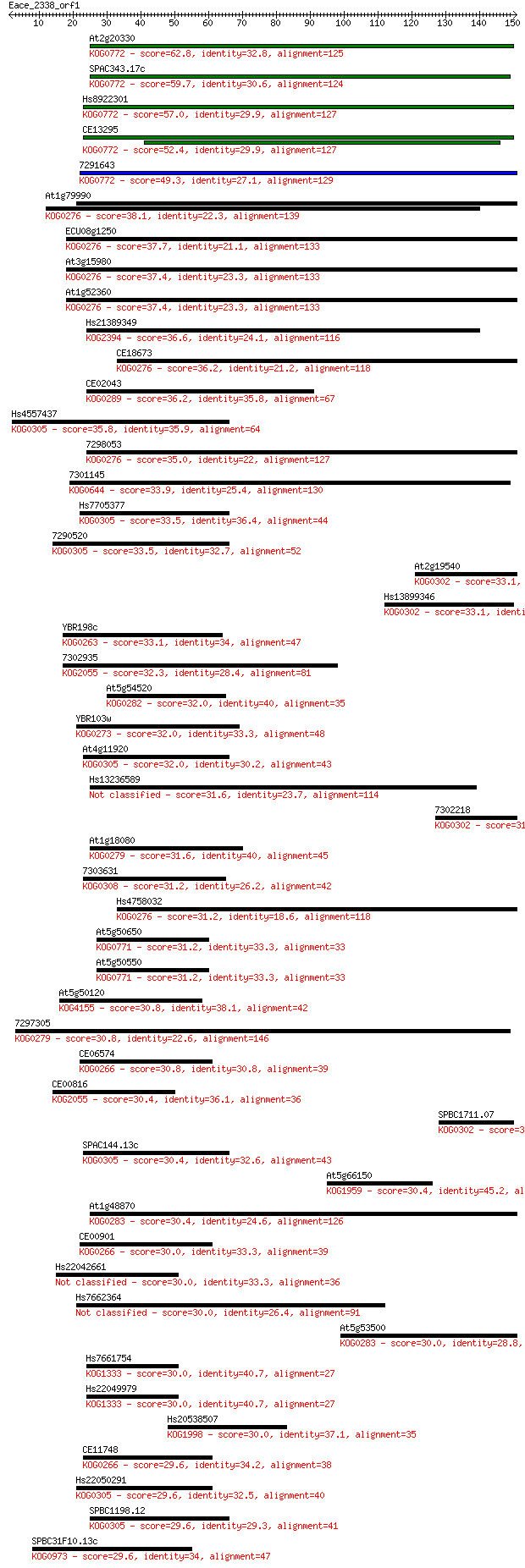
Sequences producing significant alignments: (Bits) Value
At2g20330 62.8 2e-10
SPAC343.17c 59.7 2e-09
Hs8922301 57.0 1e-08
CE13295 52.4 3e-07
7291643 49.3 3e-06
At1g79990 38.1 0.006
ECU08g1250 37.7 0.008
At3g15980 37.4 0.009
At1g52360 37.4 0.011
Hs21389349 36.6 0.018
CE18673 36.2 0.022
CE02043 36.2 0.023
Hs4557437 35.8 0.033
7298053 35.0 0.049
7301145 33.9 0.12
Hs7705377 33.5 0.14
7290520 33.5 0.17
At2g19540 33.1 0.18
Hs13899346 33.1 0.21
YBR198c 33.1 0.22
7302935 32.3 0.36
At5g54520 32.0 0.43
YBR103w 32.0 0.45
At4g11920 32.0 0.45
Hs13236589 31.6 0.56
7302218 31.6 0.62
At1g18080 31.6 0.66
7303631 31.2 0.76
Hs4758032 31.2 0.82
At5g50650 31.2 0.83
At5g50550 31.2 0.83
At5g50120 30.8 0.88
7297305 30.8 1.0
CE06574 30.8 1.1
CE00816 30.4 1.2
SPBC1711.07 30.4 1.3
SPAC144.13c 30.4 1.3
At5g66150 30.4 1.3
At1g48870 30.4 1.4
CE00901 30.0 1.6
Hs22042661 30.0 1.7
Hs7662364 30.0 1.8
At5g53500 30.0 1.8
Hs7661754 30.0 1.8
Hs22049979 30.0 1.8
Hs20538507 30.0 1.9
CE11748 29.6 2.2
Hs22050291 29.6 2.2
SPBC1198.12 29.6 2.4
SPBC31F10.13c 29.6 2.5
> At2g20330
Length=648
Score = 62.8 bits (151), Expect = 2e-10, Method: Composition-based stats.
Identities = 41/125 (32%), Positives = 64/125 (51%), Gaps = 6/125 (4%)
Query 25 VSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNMHAYRVINILERQKIKRIRFGIGVG 84
VSS A+ G R+ GS D VR+YD+ ++S + ++R I E +++ + + G
Sbjct 180 VSSLAVDSAGARVLSGSYDYTVRMYDFQGMNS--RLQSFRQIEPSEGHQVRSVSWSPTSG 237
Query 85 LKGVIGVCTGESTCYLISAAEGDLIKQTTKGDAYIRDMINTKGHTHSITDLQFNPNEKNL 144
+C S I +G + + KGD YIRD+ NTKGH +T +++P K
Sbjct 238 QF----LCVTGSAQAKIFDRDGLTLGEFMKGDMYIRDLKNTKGHICGLTCGEWHPRTKET 293
Query 145 FLTSS 149
LTSS
Sbjct 294 VLTSS 298
> SPAC343.17c
Length=576
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 38/127 (29%), Positives = 61/127 (48%), Gaps = 5/127 (3%)
Query 25 VSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNMHAYRVINILERQKIKRIRFGIG-- 82
V++ + G+R GS D + +D +S+ N H +++I+ + R+ +
Sbjct 92 VTTTTFDKNGSRFYTGSLDNTIHCWDLNGLSAT-NPHPFKIIDPTDTNADNVGRYPVSKL 150
Query 83 -VGLKGVIGVCTGESTCYLISAAEGDLIKQTTKGDAYIRDMINTKGHTHSITDLQFNPNE 141
K I S L +G LI + +KGD YIR+M NTKGH ITD + P+
Sbjct 151 SCSTKNQILALYTHSQPILYDR-DGSLIVRFSKGDQYIRNMYNTKGHIAEITDGCWQPDS 209
Query 142 KNLFLTS 148
+FLT+
Sbjct 210 SQIFLTT 216
> Hs8922301
Length=654
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 38/127 (29%), Positives = 63/127 (49%), Gaps = 6/127 (4%)
Query 23 EAVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNMHAYRVINILERQKIKRIRFGIG 82
+ VS+ L G R+ G D V+ +D+ + + + A+R + E +IK +++
Sbjct 183 KTVSALGLDPSGARLVTGGYDYDVKFWDFAGMDA--SFKAFRSLQPCECHQIKSLQYS-- 238
Query 83 VGLKGVIGVCTGESTCYLISAAEGDLIKQTTKGDAYIRDMINTKGHTHSITDLQFNPNEK 142
+I V +G S +I +G + + KGD YI DM NTKGHT + ++P K
Sbjct 239 -NTGDMILVVSGSSQAKVIDR-DGFEVMECIKGDQYIVDMANTKGHTAMLHTGSWHPKIK 296
Query 143 NLFLTSS 149
F+T S
Sbjct 297 GEFMTCS 303
> CE13295
Length=620
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 38/127 (29%), Positives = 63/127 (49%), Gaps = 6/127 (4%)
Query 23 EAVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNMHAYRVINILERQKIKRIRFGIG 82
+A+S+ + G R A G D V+L+D+ + +M + + E I + F
Sbjct 124 QAISALRVEPPGVRFASGGLDYYVKLFDFQKMDM--SMRYDKELLPAESHVINSLAFSPN 181
Query 83 VGLKGVIGVCTGESTCYLISAAEGDLIKQTTKGDAYIRDMINTKGHTHSITDLQFNPNEK 142
+ V +GE+ L+ A G +T +GD YI D+ TKGHT ++ ++FNP K
Sbjct 182 ---GETLVVASGEAIIRLLDRA-GKQWSETVRGDQYIIDLNITKGHTATVNCVEFNPLNK 237
Query 143 NLFLTSS 149
N F++ S
Sbjct 238 NEFISCS 244
Score = 31.2 bits (69), Expect = 0.75, Method: Compositional matrix adjust.
Identities = 30/107 (28%), Positives = 46/107 (42%), Gaps = 11/107 (10%)
Query 41 SEDGVVRLYDYGAVSSLKNMHAYRVINILERQKIKRIRFGIGVGLKGVIGVCTGESTCYL 100
S+DG +RL+ N+ ++VI + K I+ G + VCT
Sbjct 244 SDDGSLRLW---------NLDDHKVITKCINKHRKVIKTKGANGKRVSPQVCTFSPDGKW 294
Query 101 ISAAEGDLIKQTTK-GDAYIR-DMINTKGHTHSITDLQFNPNEKNLF 145
I+A D Q K G Y+ + + K H SIT + F+P+ K L
Sbjct 295 IAAGCDDGSVQAWKYGSQYVNVNYLVRKAHNGSITSIAFSPDSKRLL 341
> 7291643
Length=655
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 35/129 (27%), Positives = 64/129 (49%), Gaps = 6/129 (4%)
Query 22 AEAVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNMHAYRVINILERQKIKRIRFGI 81
+ AV + A G R+ GS D + +D+ + S +M ++R + E I+ +++ +
Sbjct 182 SRAVLALAGDPSGARLVSGSIDYDMCFWDFAGMDS--SMRSFRQLQPCENHPIRSLQYSV 239
Query 82 GVGLKGVIGVCTGESTCYLISAAEGDLIKQTTKGDAYIRDMINTKGHTHSITDLQFNPNE 141
+ I V +G + ++ +G + KGD YI DM TKGH +T ++P
Sbjct 240 TGDM---ILVISGNAQAKVLDR-DGFEKLECCKGDQYISDMSRTKGHVAQLTSGCWHPFN 295
Query 142 KNLFLTSSL 150
+ FLT++L
Sbjct 296 REQFLTAAL 304
> At1g79990
Length=913
Score = 38.1 bits (87), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 30/130 (23%), Positives = 54/130 (41%), Gaps = 22/130 (16%)
Query 21 TAEAVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNMHAYRVINILERQKIKRIRFG 80
T V S R + G++D +R+Y+Y + +K A+ IR
Sbjct 49 TELPVRSAKFIARKQWVVAGADDMFIRVYNYNTMDKIKVFEAH----------ADYIR-- 96
Query 81 IGVGLKGVIGVCTGESTCYLISAAEGDLIKQTTKGDAYIRDMINTKGHTHSITDLQFNPN 140
V + Y++S+++ LIK ++ I +GH+H + + FNP
Sbjct 97 ---------CVAVHPTLPYVLSSSDDMLIKLWDWEKGWLCTQI-FEGHSHYVMQVTFNPK 146
Query 141 EKNLFLTSSL 150
+ N F ++SL
Sbjct 147 DTNTFASASL 156
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 32/154 (20%), Positives = 65/154 (42%), Gaps = 29/154 (18%)
Query 12 LGRFSFIEDTAEAVSSFALSQRGNRIAIGSEDGVVRLYDY---GAVSSLKNMHAYRVINI 68
+ + E A+ + A+ + S+D +++L+D+ + + H++ V+ +
Sbjct 82 MDKIKVFEAHADYIRCVAVHPTLPYVLSSSDDMLIKLWDWEKGWLCTQIFEGHSHYVMQV 141
Query 69 --------------LERQ-KIKRI-----RFGIGVGLKGV--IGVCTGESTCYLISAAEG 106
L+R KI + F + LKGV + TG YLI+ ++
Sbjct 142 TFNPKDTNTFASASLDRTIKIWNLGSPDPNFTLDAHLKGVNCVDYFTGGDKPYLITGSDD 201
Query 107 DLIKQTTKGDAYIRDMINT-KGHTHSITDLQFNP 139
K D + + T +GHTH+++ + F+P
Sbjct 202 HTAK---VWDYQTKSCVQTLEGHTHNVSAVSFHP 232
> ECU08g1250
Length=759
Score = 37.7 bits (86), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 28/133 (21%), Positives = 57/133 (42%), Gaps = 22/133 (16%)
Query 18 IEDTAEAVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNMHAYRVINILERQKIKRI 77
I + E + + + R + + +GS+DG V +Y+ G +K HA+ I++I
Sbjct 89 IHVSNEPIRTCVILGRMDWLLVGSDDGNVSIYELGKYRKVKTFHAHDDF-------IRKI 141
Query 78 RFGIGVGLKGVIGVCTGESTCYLISAAEGDLIKQTTKGDAYIRDMINTKGHTHSITDLQF 137
+ +L ++ + L +G+ + + GH H + D+ F
Sbjct 142 E-------------SHPQDPSFLTASDDATLKMWIYQGE--VSQAMTYTGHEHFVMDVCF 186
Query 138 NPNEKNLFLTSSL 150
PN+ + F++ SL
Sbjct 187 YPNDASKFVSCSL 199
> At3g15980
Length=921
Score = 37.4 bits (85), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 31/133 (23%), Positives = 55/133 (41%), Gaps = 22/133 (16%)
Query 18 IEDTAEAVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNMHAYRVINILERQKIKRI 77
E T V S R + G++D +R+Y+Y + +K A+ I
Sbjct 56 FEVTELPVRSAKFIPRKQWVVAGADDMYIRVYNYNTMDKVKVFEAHS----------DYI 105
Query 78 RFGIGVGLKGVIGVCTGESTCYLISAAEGDLIKQTTKGDAYIRDMINTKGHTHSITDLQF 137
R V + Y++S+++ LIK + + I +GH+H + + F
Sbjct 106 R-----------CVAVHPTLPYVLSSSDDMLIKLWDWENGWACTQI-FEGHSHYVMQVVF 153
Query 138 NPNEKNLFLTSSL 150
NP + N F ++SL
Sbjct 154 NPKDTNTFASASL 166
> At1g52360
Length=926
Score = 37.4 bits (85), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 31/133 (23%), Positives = 54/133 (40%), Gaps = 22/133 (16%)
Query 18 IEDTAEAVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNMHAYRVINILERQKIKRI 77
E T V S R + G++D +R+Y+Y + +K A+ I
Sbjct 53 FEVTELPVRSAKFVARKQWVVAGADDMYIRVYNYNTMDKVKVFEAHS----------DYI 102
Query 78 RFGIGVGLKGVIGVCTGESTCYLISAAEGDLIKQTTKGDAYIRDMINTKGHTHSITDLQF 137
R V + Y++S+++ LIK + I +GH+H + + F
Sbjct 103 R-----------CVAVHPTLPYVLSSSDDMLIKLWDWEKGWACTQI-FEGHSHYVMQVTF 150
Query 138 NPNEKNLFLTSSL 150
NP + N F ++SL
Sbjct 151 NPKDTNTFASASL 163
> Hs21389349
Length=569
Score = 36.6 bits (83), Expect = 0.018, Method: Composition-based stats.
Identities = 28/116 (24%), Positives = 51/116 (43%), Gaps = 23/116 (19%)
Query 24 AVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNMHAYRVINILERQKIKRIRFGIGV 83
A++ FA S G +A S+DG +R++++ +V M +Y FG
Sbjct 222 ALNEFAFSPDGKFLACVSQDGFLRVFNFDSVELHGTMKSY---------------FG--- 263
Query 84 GLKGVIGVCTGESTCYLISAAEGDLIKQTTKGDAYIRDMINTKGHTHSITDLQFNP 139
G++ VC Y+++ E DL+ + D R + GH ++ + F+P
Sbjct 264 ---GLLCVCWSPDGKYIVTGGEDDLVTVWSFVDC--RVIARGHGHKSWVSVVAFDP 314
> CE18673
Length=1000
Score = 36.2 bits (82), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 25/118 (21%), Positives = 53/118 (44%), Gaps = 22/118 (18%)
Query 33 RGNRIAIGSEDGVVRLYDYGAVSSLKNMHAYRVINILERQKIKRIRFGIGVGLKGVIGVC 92
R + + GS+D +R+++Y ++L+ +H + + R +
Sbjct 68 RKSWVVTGSDDMHIRVFNY---NTLERVHQFEAHSDYLR------------------SLV 106
Query 93 TGESTCYLISAAEGDLIKQTTKGDAYIRDMINTKGHTHSITDLQFNPNEKNLFLTSSL 150
+ Y+IS+++ L+K + + + +GHTH + + NP + N F T+SL
Sbjct 107 VHPTLPYVISSSDDMLVKMWDWDNKWAMKQ-SFEGHTHYVMQIAINPKDNNTFATASL 163
> CE02043
Length=509
Score = 36.2 bits (82), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 35/67 (52%), Gaps = 6/67 (8%)
Query 24 AVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNMHAYRVINILERQKIKRIRFGIGV 83
AV S A S+ G +A GSEDG V+L+D + LKN+ + E+Q I + F +
Sbjct 399 AVRSIAFSENGYYLATGSEDGEVKLWD---LRKLKNLKTFAN---EEKQPINSLSFDMTG 452
Query 84 GLKGVIG 90
G+ G
Sbjct 453 TFLGIGG 459
> Hs4557437
Length=499
Score = 35.8 bits (81), Expect = 0.033, Method: Composition-based stats.
Identities = 23/66 (34%), Positives = 36/66 (54%), Gaps = 6/66 (9%)
Query 2 AAAAKDELPLLGRFSFIEDTAEAVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNM- 60
+A++ D L LL +E E +SS A + GN +A+G+ V+L+D L+NM
Sbjct 210 SASSGDILQLLQ----MEQPGEYISSVAWIKEGNYLAVGTSSAEVQLWDVQQQKRLRNMT 265
Query 61 -HAYRV 65
H+ RV
Sbjct 266 SHSARV 271
> 7298053
Length=860
Score = 35.0 bits (79), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 28/131 (21%), Positives = 53/131 (40%), Gaps = 30/131 (22%)
Query 24 AVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNMHAY----RVINILERQKIKRIRF 79
V S R N I GS+D +R+++Y + + + A+ R I + Q +
Sbjct 30 PVRSARFVARKNWILTGSDDMQIRVFNYNTLEKVHSFEAHSDYLRCIAVHPTQPL----- 84
Query 80 GIGVGLKGVIGVCTGESTCYLISAAEGDLIKQTTKGDAYIRDMINTKGHTHSITDLQFNP 139
++++++ LIK + + +GHTH + + FNP
Sbjct 85 --------------------VLTSSDDMLIKLWNWEKMWACQRV-FEGHTHYVMQIVFNP 123
Query 140 NEKNLFLTSSL 150
+ N F ++SL
Sbjct 124 KDNNTFASASL 134
> 7301145
Length=1225
Score = 33.9 bits (76), Expect = 0.12, Method: Composition-based stats.
Identities = 33/132 (25%), Positives = 59/132 (44%), Gaps = 13/132 (9%)
Query 19 EDTAEAVSSFALSQRGNRIAIGSEDGVVRLYDYGAVS--SLKNMHAYRVINILERQKIKR 76
E +AV S S RG R GS+DG ++ + + S K R+ + E ++ KR
Sbjct 366 EAYTDAVDSVQWSHRGLRFISGSKDGTAHIWTFESQQWKSSKLCMTERLASCPEPEEGKR 425
Query 77 IRFGIGVGLKGVIGVCTGESTCYLISAAEGDLIKQTTKGDAYIRDMINTKGHTHSITDLQ 136
++ V V S Y+I+A IK A + ++ +GH + L+
Sbjct 426 LK---------VTMVAWDASDRYVITAVNDFTIKIWDSKSAKLHRVL--RGHKDELYVLE 474
Query 137 FNPNEKNLFLTS 148
NP ++++ L++
Sbjct 475 SNPRDEHVLLSA 486
> Hs7705377
Length=496
Score = 33.5 bits (75), Expect = 0.14, Method: Composition-based stats.
Identities = 16/46 (34%), Positives = 27/46 (58%), Gaps = 2/46 (4%)
Query 22 AEAVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNM--HAYRV 65
++V+S S+RGN +A+G+ G V+++D A L + H RV
Sbjct 229 GDSVTSVGWSERGNLVAVGTHKGFVQIWDAAAGKKLSMLEGHTARV 274
> 7290520
Length=478
Score = 33.5 bits (75), Expect = 0.17, Method: Composition-based stats.
Identities = 17/54 (31%), Positives = 29/54 (53%), Gaps = 2/54 (3%)
Query 14 RFSFIEDTAEAVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLK--NMHAYRV 65
R + A V+S + ++RGN +A+G+ G V ++D A + N H+ RV
Sbjct 206 RLCDLSPDANTVTSVSWNERGNTVAVGTHHGYVTVWDVAANKQINKLNGHSARV 259
> At2g19540
Length=469
Score = 33.1 bits (74), Expect = 0.18, Method: Composition-based stats.
Identities = 14/30 (46%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 121 DMINTKGHTHSITDLQFNPNEKNLFLTSSL 150
D I GHT S+ DLQ++P E+N+F + S+
Sbjct 261 DPIPFAGHTASVEDLQWSPAEENVFASCSV 290
> Hs13899346
Length=446
Score = 33.1 bits (74), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 112 TTKGDAYIRDMINTKGHTHSITDLQFNPNEKNLFLTSS 149
T G ++ D GHT S+ DLQ++P E +F + S
Sbjct 244 PTDGGSWHVDQRPFVGHTRSVEDLQWSPTENTVFASCS 281
> YBR198c
Length=798
Score = 33.1 bits (74), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 26/47 (55%), Gaps = 1/47 (2%)
Query 17 FIEDTAEAVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNMHAY 63
F+ TA +S A+ G ++ GSEDG++ ++D G LK M +
Sbjct 647 FLGHTAPVIS-IAVCPDGRWLSTGSEDGIINVWDIGTGKRLKQMRGH 692
> 7302935
Length=506
Score = 32.3 bits (72), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 23/82 (28%), Positives = 40/82 (48%), Gaps = 6/82 (7%)
Query 17 FIEDTAEAVSSFALSQRGNRIAIGSEDGVVRLYDYGAV-SSLKNMHAYRVINILERQKIK 75
F++D S LS +A GS++GVV +YDY ++ +S R +N+ R I
Sbjct 371 FMDDGCIHGESIQLSPNQRLLATGSQEGVVNIYDYESIFASKAPQPEKRFMNL--RTAIT 428
Query 76 RIRFGIGVGLKGVIGVCTGEST 97
++F L + +C+ E+
Sbjct 429 DLQFNHSSEL---LAMCSSEAP 447
> At5g54520
Length=404
Score = 32.0 bits (71), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 30 LSQRGNRIAIGSEDGVVRLYDYGAVSSLKNMHAYR 64
S G +A GS DG V +YDY + + +K + AY
Sbjct 341 FSPDGETLASGSSDGSVYMYDYKSTALIKKLKAYE 375
> YBR103w
Length=535
Score = 32.0 bits (71), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 28/49 (57%), Gaps = 1/49 (2%)
Query 21 TAEAVSSFALSQRGNRIAIGSEDGVVRLYD-YGAVSSLKNMHAYRVINI 68
T V+ A S GN I G E+G +RL++ GA+ ++ N H ++++
Sbjct 219 TTNQVTCLAWSHDGNSIVTGVENGELRLWNKTGALLNVLNFHRAPIVSV 267
> At4g11920
Length=482
Score = 32.0 bits (71), Expect = 0.45, Method: Composition-based stats.
Identities = 13/43 (30%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 23 EAVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNMHAYRV 65
E V S + RG +AIG+ G V+++D +++ M +R+
Sbjct 224 ETVCSVGWALRGTHLAIGTSSGTVQIWDVLRCKNIRTMEGHRL 266
> Hs13236589
Length=317
Score = 31.6 bits (70), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 27/122 (22%), Positives = 48/122 (39%), Gaps = 14/122 (11%)
Query 25 VSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNMHAYRVINILERQKIKRIRFGIGVG 84
+ AL +R + G EDG VRL+D L+ + I + + ++ R G +G
Sbjct 147 IHCLALRERSPEVLSGGEDGAVRLWD------LRTAKEVQTIEVYKHEECSRPHNGRWIG 200
Query 85 LKGV---IGVCTGEST---CYLISAAEGDL--IKQTTKGDAYIRDMINTKGHTHSITDLQ 136
VC G +L S+ + I+ K + +D+I + G + Q
Sbjct 201 CLATDSDWMVCGGGPALTLWHLRSSTPTTIFPIRAPQKHVTFYQDLILSAGQGRCVNQWQ 260
Query 137 FN 138
+
Sbjct 261 LS 262
> 7302218
Length=422
Score = 31.6 bits (70), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 10/24 (41%), Positives = 19/24 (79%), Gaps = 0/24 (0%)
Query 127 GHTHSITDLQFNPNEKNLFLTSSL 150
GH+ S+ DLQ++PNE+++ + S+
Sbjct 233 GHSQSVEDLQWSPNERSVLASCSV 256
> At1g18080
Length=327
Score = 31.6 bits (70), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 18/45 (40%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 25 VSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNMHAYRVINIL 69
VS+ A+S G+ A G +DGVV L+D L ++ A VI+ L
Sbjct 197 VSTVAVSPDGSLCASGGKDGVVLLWDLAEGKKLYSLEANSVIHAL 241
> 7303631
Length=680
Score = 31.2 bits (69), Expect = 0.76, Method: Composition-based stats.
Identities = 11/42 (26%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 23 EAVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNMHAYR 64
E V +S GN++ GS DG +++++ G ++ +H ++
Sbjct 209 ENVRCLVVSPDGNQVVSGSSDGTIKVWNLGQQRCVQTIHVHK 250
> Hs4758032
Length=906
Score = 31.2 bits (69), Expect = 0.82, Method: Compositional matrix adjust.
Identities = 22/118 (18%), Positives = 52/118 (44%), Gaps = 22/118 (18%)
Query 33 RGNRIAIGSEDGVVRLYDYGAVSSLKNMHAYRVINILERQKIKRIRFGIGVGLKGVIGVC 92
R N + G++D +R+++Y ++L+ +H + + R +
Sbjct 68 RKNWVVTGADDMQIRVFNY---NTLERVHMFEAHSDYIR------------------CIA 106
Query 93 TGESTCYLISAAEGDLIKQTTKGDAYIRDMINTKGHTHSITDLQFNPNEKNLFLTSSL 150
+ +++++++ LIK + + +GHTH + + NP + N F ++SL
Sbjct 107 VHPTQPFILTSSDDMLIKLWDWDKKWSCSQV-FEGHTHYVMQIVINPKDNNQFASASL 163
> At5g50650
Length=383
Score = 31.2 bits (69), Expect = 0.83, Method: Compositional matrix adjust.
Identities = 11/33 (33%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 27 SFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKN 59
S + +Q G +A G+EDG +R++++ ++ +L N
Sbjct 147 SLSFNQDGTVLATGAEDGTLRVFEWPSMKTLLN 179
> At5g50550
Length=383
Score = 31.2 bits (69), Expect = 0.83, Method: Compositional matrix adjust.
Identities = 11/33 (33%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 27 SFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKN 59
S + +Q G +A G+EDG +R++++ ++ +L N
Sbjct 147 SLSFNQDGTVLATGAEDGTLRVFEWPSMKTLLN 179
> At5g50120
Length=388
Score = 30.8 bits (68), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 25/42 (59%), Gaps = 2/42 (4%)
Query 16 SFIEDTAEAVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSL 57
+ + TA +SS L+ G R+ GS DGVVRL++ + +L
Sbjct 32 TLVCHTASYISSLTLA--GKRLYTGSNDGVVRLWNANTLETL 71
> 7297305
Length=318
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 33/158 (20%), Positives = 59/158 (37%), Gaps = 34/158 (21%)
Query 3 AAAKDELPLLGRFSFIEDT------------AEAVSSFALSQRGNRIAIGSEDGVVRLYD 50
+A++D+ ++ + + EDT + +S LS GN GS D +RL+D
Sbjct 33 SASRDKTLIVWKLTRDEDTNYGYPQKRLYGHSHFISDVVLSSDGNYALSGSWDQTLRLWD 92
Query 51 YGAVSSLKNMHAYRVINILERQKIKRIRFGIGVGLKGVIGVCTGESTCYLISAAEGDLIK 110
A + + + K V+ V ++S + IK
Sbjct 93 LAAGKTTRRFEGH---------------------TKDVLSVAFSADNRQIVSGSRDKTIK 131
Query 111 QTTKGDAYIRDMINTKGHTHSITDLQFNPNEKNLFLTS 148
A + I GHT ++ ++F+PN N + S
Sbjct 132 LWNTL-AECKFTIQEDGHTDWVSCVRFSPNHSNPIIVS 168
> CE06574
Length=501
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 12/39 (30%), Positives = 24/39 (61%), Gaps = 0/39 (0%)
Query 22 AEAVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNM 60
++ ++S + + GN +A S DG +R++D + S LK +
Sbjct 339 SDPITSISYNHDGNTMATSSYDGCIRVWDAASGSCLKTL 377
> CE00816
Length=429
Score = 30.4 bits (67), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 14 RFSFIEDTAEAVSSFALSQRGNRIAIGSEDGVVRLY 49
+ +F +D A ++ A+SQ G+ A GS+ G+V +Y
Sbjct 291 QHTFTDDGAVHGTTLAISQHGDYFATGSDTGIVNVY 326
> SPBC1711.07
Length=480
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 11/22 (50%), Positives = 18/22 (81%), Gaps = 0/22 (0%)
Query 128 HTHSITDLQFNPNEKNLFLTSS 149
HT ++ DLQ++P+EKN+F + S
Sbjct 290 HTAAVEDLQWSPSEKNVFSSCS 311
> SPAC144.13c
Length=556
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 14/45 (31%), Positives = 23/45 (51%), Gaps = 2/45 (4%)
Query 23 EAVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNM--HAYRV 65
+ V+S QRG +A+G+ +G V ++D + M H RV
Sbjct 292 DTVTSLRWVQRGTHLAVGTHNGSVEIWDAATCKKTRTMSGHTERV 336
> At5g66150
Length=1047
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 19/31 (61%), Gaps = 5/31 (16%)
Query 95 ESTCYLISAAEGDLIKQTTKGDAYIRDMINT 125
E+TC+ I D+I QTTKG +I+ NT
Sbjct 144 EATCHYI-----DMIDQTTKGHRFIKQQFNT 169
> At1g48870
Length=593
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 31/136 (22%), Positives = 55/136 (40%), Gaps = 13/136 (9%)
Query 25 VSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLK----------NMHAYRVINILERQKI 74
+ + S G +A G EDGVV+++ SL N A V+ + I
Sbjct 201 IWTLKFSPDGKYLATGGEDGVVKIWRITLSDSLLASFLRQQEPINQQAALVLFPQKAFHI 260
Query 75 KRIRFGIGVGLKGVIGVCTGESTCYLISAAEGDLIKQTTKGDAYIRDMINTKGHTHSITD 134
+ F G G + + L+SA++ ++ G ++ H + +T
Sbjct 261 EETPFQELYGHTGDVLDLAWSDSNLLLSASKDKTVRLWRTG---CDQCLHVFHHNNYVTC 317
Query 135 LQFNPNEKNLFLTSSL 150
++FNP KN F + S+
Sbjct 318 VEFNPVNKNNFASGSI 333
> CE00901
Length=376
Score = 30.0 bits (66), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 13/39 (33%), Positives = 25/39 (64%), Gaps = 0/39 (0%)
Query 22 AEAVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNM 60
++ VS+ + ++ G+ IA GS DG+VR++D +K +
Sbjct 213 SDPVSAVSFNRDGSLIASGSYDGLVRIWDTANGQCIKTL 251
> Hs22042661
Length=2266
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 12/36 (33%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 15 FSFIEDTAEAVSSFALSQRGNRIAIGSEDGVVRLYD 50
F + T AVS+ + R N++A+G DG + L++
Sbjct 213 FQLVSPTGTAVSTLSYISRTNQLAVGFSDGYLALWN 248
> Hs7662364
Length=919
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 24/92 (26%), Positives = 39/92 (42%), Gaps = 13/92 (14%)
Query 21 TAEAVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNMHAYRVINILERQKIKRIRFG 80
T AV+S A+ G +A G ED LYD +++ H + ++ +RF
Sbjct 799 TGSAVASVAVDPSGRLLATGQEDSSCMLYDIRGGRMVQSYHPH-------SSDVRSVRFS 851
Query 81 IGVGLKGVIGVCTGESTCYL-ISAAEGDLIKQ 111
G + TG + ++ +GDL KQ
Sbjct 852 PGAHY-----LLTGSYDMKIKVTDLQGDLTKQ 878
> At5g53500
Length=654
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 15/52 (28%), Positives = 26/52 (50%), Gaps = 3/52 (5%)
Query 99 YLISAAEGDLIKQTTKGDAYIRDMINTKGHTHSITDLQFNPNEKNLFLTSSL 150
YL+SA+ ++ G D + H +T +QFNP +N F++ S+
Sbjct 341 YLLSASMDKTVRLWKVGS---NDCLGVFAHNSYVTSVQFNPVNENYFMSGSI 389
> Hs7661754
Length=741
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 11/27 (40%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 24 AVSSFALSQRGNRIAIGSEDGVVRLYD 50
A++ A + GN + G+ DGV+RL+D
Sbjct 559 AINCTAFNHNGNLLVTGAADGVIRLFD 585
> Hs22049979
Length=747
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 11/27 (40%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 24 AVSSFALSQRGNRIAIGSEDGVVRLYD 50
A++ A + GN + G+ DGV+RL+D
Sbjct 564 AINCTAFNHNGNLLVTGAADGVIRLFD 590
> Hs20538507
Length=988
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 23/35 (65%), Gaps = 0/35 (0%)
Query 48 LYDYGAVSSLKNMHAYRVINILERQKIKRIRFGIG 82
LYDY ++S ++ M A NI+E+Q+++ F +G
Sbjct 877 LYDYQSLSWIRKMEASYYDNIMEQQRLEPEFFRVG 911
> CE11748
Length=395
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 23 EAVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNM 60
+ VSS ++ G +A GS DG+VR++D + +K +
Sbjct 233 DPVSSVCFNRDGAYLASGSYDGIVRIWDSTTGTCVKTL 270
> Hs22050291
Length=372
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 13/40 (32%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 21 TAEAVSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNM 60
T +SS + + G +A+G+ +G V+L+D L+NM
Sbjct 272 TCNYISSVSWIKEGTCLAVGTSEGEVQLWDVVTKKRLRNM 311
> SPBC1198.12
Length=421
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 12/43 (27%), Positives = 29/43 (67%), Gaps = 2/43 (4%)
Query 25 VSSFALSQRGNRIAIGSEDGVVRLYDYGAVSSLKNM--HAYRV 65
V+S + +G ++A+G++ GV+ ++D + S++++ H+ RV
Sbjct 169 VTSVLWTGKGTQLAVGTDSGVIYIWDIESTKSVRSLKGHSERV 211
> SPBC31F10.13c
Length=932
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 27/49 (55%), Gaps = 5/49 (10%)
Query 8 ELPLLGRFSFIEDTAEAVSSFALS--QRGNRIAIGSEDGVVRLYDYGAV 54
++P LG F +D +S F++ G+RIA G DG +R++ A+
Sbjct 5 KIPWLGHF---DDRGHRLSIFSIHIHPDGSRIATGGLDGTIRIWSTEAI 50
Lambda K H
0.319 0.136 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1852391706
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40