bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2326_orf1
Length=102
Score E
Sequences producing significant alignments: (Bits) Value
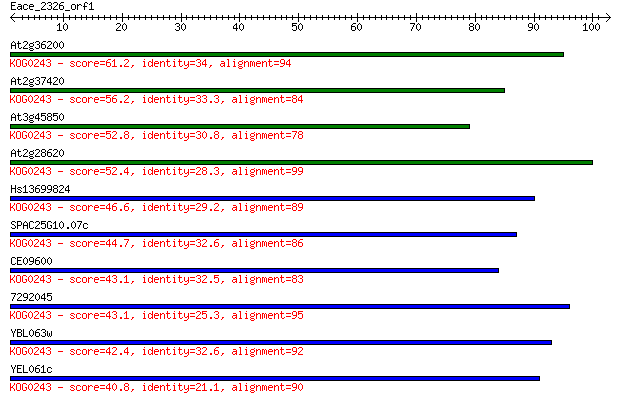
At2g36200 61.2 4e-10
At2g37420 56.2 1e-08
At3g45850 52.8 2e-07
At2g28620 52.4 2e-07
Hs13699824 46.6 1e-05
SPAC25G10.07c 44.7 4e-05
CE09600 43.1 1e-04
7292045 43.1 1e-04
YBL063w 42.4 2e-04
YEL061c 40.8 7e-04
> At2g36200
Length=1056
Score = 61.2 bits (147), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 32/94 (34%), Positives = 56/94 (59%), Gaps = 0/94 (0%)
Query 1 EETLCTLDYAYRAKSITTRPVPTLRRSADLLLTQLLLENSQLRLLLQQQRERDGVFLPLD 60
EETL TLDYA+RAK+I +P + L+ L E +L+ + RE++GV++P +
Sbjct 344 EETLSTLDYAHRAKNIRNKPEVNQKMMKSTLIKDLYGEIERLKAEVYASREKNGVYMPKE 403
Query 61 MFQQQQQQQQQQQQQIQQQAAQLHALQQQTEQQQ 94
+ Q++ +++ +QI+Q Q+ Q+Q E+ Q
Sbjct 404 RYYQEESERKVMAEQIEQMGGQIENYQKQLEELQ 437
> At2g37420
Length=1022
Score = 56.2 bits (134), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 28/84 (33%), Positives = 55/84 (65%), Gaps = 0/84 (0%)
Query 1 EETLCTLDYAYRAKSITTRPVPTLRRSADLLLTQLLLENSQLRLLLQQQRERDGVFLPLD 60
EETL TLDYAYRAK+I +P + S +LL L LE +++ ++ R+++GV++ +
Sbjct 375 EETLSTLDYAYRAKNIKNKPEANQKLSKAVLLKDLYLELERMKEDVRAARDKNGVYIAHE 434
Query 61 MFQQQQQQQQQQQQQIQQQAAQLH 84
+ Q++ +++ + ++I+Q +L+
Sbjct 435 RYTQEEVEKKARIERIEQLENELN 458
> At3g45850
Length=1058
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 48/78 (61%), Gaps = 0/78 (0%)
Query 1 EETLCTLDYAYRAKSITTRPVPTLRRSADLLLTQLLLENSQLRLLLQQQRERDGVFLPLD 60
EETL TLDYA+RAK+I +P + ++ L E +L+ + RE++G+++P D
Sbjct 375 EETLSTLDYAHRAKNIKNKPEINQKMMKSAVMKDLYSEIDRLKQEVYAAREKNGIYIPKD 434
Query 61 MFQQQQQQQQQQQQQIQQ 78
+ Q++ +++ ++I++
Sbjct 435 RYIQEEAEKKAMAEKIER 452
> At2g28620
Length=1076
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/99 (28%), Positives = 54/99 (54%), Gaps = 0/99 (0%)
Query 1 EETLCTLDYAYRAKSITTRPVPTLRRSADLLLTQLLLENSQLRLLLQQQRERDGVFLPLD 60
EETL TLDYA+RAK I +P + ++ L E +L+ + RE++G+++P +
Sbjct 377 EETLSTLDYAHRAKHIKNKPEVNQKMMKSAIMKDLYSEIERLKQEVYAAREKNGIYIPKE 436
Query 61 MFQQQQQQQQQQQQQIQQQAAQLHALQQQTEQQQQTINT 99
+ Q++ +++ +I+Q + A +Q Q+ N+
Sbjct 437 RYTQEEAEKKAMADKIEQMEVEGEAKDKQIIDLQELYNS 475
> Hs13699824
Length=1056
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 52/89 (58%), Gaps = 0/89 (0%)
Query 1 EETLCTLDYAYRAKSITTRPVPTLRRSADLLLTQLLLENSQLRLLLQQQRERDGVFLPLD 60
EETL TL+YA+RAK+I +P + + L+ + E +L+ L RE++GV++ +
Sbjct 344 EETLSTLEYAHRAKNILNKPEVNQKLTKKALIKEYTEEIERLKRDLAAAREKNGVYISEE 403
Query 61 MFQQQQQQQQQQQQQIQQQAAQLHALQQQ 89
F+ + Q++QI + ++ A++++
Sbjct 404 NFRVMSGKLTVQEEQIVELIEKIGAVEEE 432
> SPAC25G10.07c
Length=1085
Score = 44.7 bits (104), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 28/86 (32%), Positives = 48/86 (55%), Gaps = 0/86 (0%)
Query 1 EETLCTLDYAYRAKSITTRPVPTLRRSADLLLTQLLLENSQLRLLLQQQRERDGVFLPLD 60
EET+ TL+YA RAKSI +P +L+ L+L+ +L+ L R+++GV+L
Sbjct 406 EETISTLEYAARAKSIRNKPQNNQLVFRKVLIKDLVLDIERLKNDLNATRKKNGVYLAES 465
Query 61 MFQQQQQQQQQQQQQIQQQAAQLHAL 86
+++ + Q + Q+QA +L L
Sbjct 466 TYKELMDRVQNKDLLCQEQARKLEVL 491
> CE09600
Length=958
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 46/86 (53%), Gaps = 3/86 (3%)
Query 1 EETLCTLDYAYRAKSITTRPVPTLRRSADLLLTQLLLENSQLRLLLQQQRERDGVFL--- 57
EE+ TL+YA RA +I +PV + S +L + E +LR L+ RE++GV +
Sbjct 330 EESQSTLEYAMRAANIKNKPVCNTKLSKKTILKEYSDEIEKLRRDLRAAREKNGVIISQE 389
Query 58 PLDMFQQQQQQQQQQQQQIQQQAAQL 83
D FQ+ ++ Q+ +Q + +L
Sbjct 390 SHDEFQKNSEKVQELEQHLDNAVDRL 415
> 7292045
Length=1048
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 55/95 (57%), Gaps = 0/95 (0%)
Query 1 EETLCTLDYAYRAKSITTRPVPTLRRSADLLLTQLLLENSQLRLLLQQQRERDGVFLPLD 60
EETL TL+YA+RAK+I +P + + +L + E +L+ L R+++G++L +
Sbjct 341 EETLSTLEYAHRAKNIQNKPEVNQKLTKKTVLKEYTEEIDKLKRDLMAARDKNGIYLAEE 400
Query 61 MFQQQQQQQQQQQQQIQQQAAQLHALQQQTEQQQQ 95
+ + + + Q +++ ++ L AL+ + + +++
Sbjct 401 TYGEITLKLESQNRELNEKMLLLKALKDELQNKEK 435
> YBL063w
Length=1111
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 30/95 (31%), Positives = 47/95 (49%), Gaps = 10/95 (10%)
Query 1 EETLCTLDYAYRAKSITTRPVPTLRRSADLLLTQLLLENSQLRLLLQQQRERDGVFLP-- 58
EET TL+YA RAKSI P S D L + E +LR L+ R + G+F+
Sbjct 395 EETASTLEYATRAKSIKNTPQVNQSLSKDTCLKDYIQEIEKLRNDLKNSRNKQGIFITQD 454
Query 59 -LDMFQQQQQQQQQQQQQIQQQAAQLHALQQQTEQ 92
LD++ + I +Q ++H L++Q ++
Sbjct 455 QLDLY-------ESNSILIDEQNLKIHNLREQIKK 482
> YEL061c
Length=1038
Score = 40.8 bits (94), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 19/90 (21%), Positives = 48/90 (53%), Gaps = 0/90 (0%)
Query 1 EETLCTLDYAYRAKSITTRPVPTLRRSADLLLTQLLLENSQLRLLLQQQRERDGVFLPLD 60
EET TL+YA +AK+I +P D+L+ + +E ++++ L + ++G+++ D
Sbjct 500 EETCSTLEYASKAKNIKNKPQLGSFIMKDILVKNITMELAKIKSDLLSTKSKEGIYMSQD 559
Query 61 MFQQQQQQQQQQQQQIQQQAAQLHALQQQT 90
++ + + ++Q+ ++ +L +
Sbjct 560 HYKNLNSDLESYKNEVQECKREIESLTSKN 589
Lambda K H
0.317 0.126 0.331
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1181107380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40