bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2312_orf3
Length=178
Score E
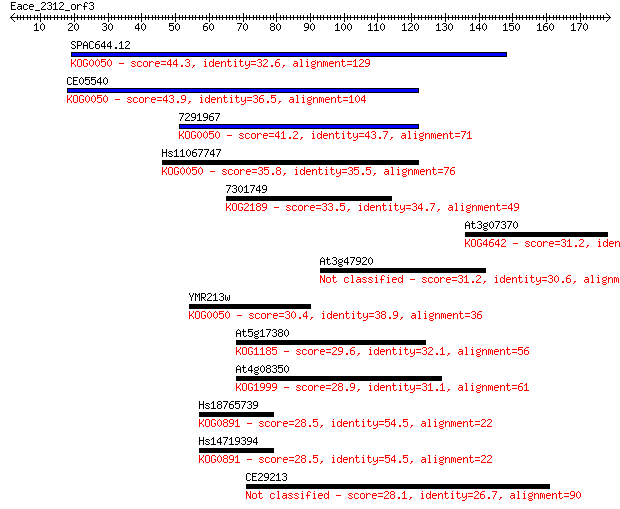
Sequences producing significant alignments: (Bits) Value
SPAC644.12 44.3 1e-04
CE05540 43.9 2e-04
7291967 41.2 0.001
Hs11067747 35.8 0.041
7301749 33.5 0.23
At3g07370 31.2 1.0
At3g47920 31.2 1.2
YMR213w 30.4 1.8
At5g17380 29.6 3.3
At4g08350 28.9 5.8
Hs18765739 28.5 8.1
Hs14719394 28.5 8.2
CE29213 28.1 8.6
> SPAC644.12
Length=757
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 42/134 (31%), Positives = 64/134 (47%), Gaps = 6/134 (4%)
Query 19 ATPLNDPSDAGS----ASEGGGMGSKAELQLARLQA-RTSLASLPAPRNDVTGQIPDDIT 73
ATP P D S A G + S+ E ++ RL+A R LA LP P+ND P
Sbjct 432 ATPFRTPRDTFSINAAAERAGRLASERENKI-RLKALRELLAKLPKPKNDYELMEPRFAD 490
Query 74 EEEKELLESEADLDMEEVERRRAQRAAEAARRRFLEQTQVMQRQLPRPILPPPKLLTSSI 133
E + E + D + ERR +R AE R ++QV+QR L RP + P+ S+
Sbjct 491 ETDVEATVGVLEEDATDRERRIQERIAEKERLAKARRSQVIQRDLIRPSVTQPEKWKRSL 550
Query 134 DAASAIASAVQQQL 147
+ A+ + +++
Sbjct 551 ENEDPTANVLLKEM 564
> CE05540
Length=755
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 38/111 (34%), Positives = 55/111 (49%), Gaps = 16/111 (14%)
Query 18 SATPLNDPSDAGSASEGGGMGSKAELQLARLQARTSLASLPAPRNDVTGQIPDDITEEEK 77
+ATP D G + KA L+ A LASLP P+ND PDD +E +
Sbjct 431 AATPFRDQMRINEEIAGSALEQKASLKRA-------LASLPTPKNDFEVVGPDD--DEVE 481
Query 78 ELLESEADLD----MEEVERR---RAQRAAEAARRRFLEQTQVMQRQLPRP 121
+E E++ D +E+ R +A+R AE R ++QV+QR LP+P
Sbjct 482 GAVEDESNQDEDGWIEDASERAENKAKRNAENRVRNMKMRSQVIQRSLPKP 532
> 7291967
Length=791
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 31/74 (41%), Positives = 47/74 (63%), Gaps = 6/74 (8%)
Query 51 RTSLASLPAPRNDVTGQIPDDITEEEKELLESEADLDMEEVERRRAQRAA--EAARRRFL 108
R L++LPAPRND +P+ +EE E +E+ ++ +E+ A+ A EA R+R L
Sbjct 457 RDGLSTLPAPRNDYEIVVPE---QEESERIETNSEPAVEDQADVDARLLAEQEARRKREL 513
Query 109 EQ-TQVMQRQLPRP 121
E+ +QV+QR LPRP
Sbjct 514 EKRSQVIQRSLPRP 527
> Hs11067747
Length=802
Score = 35.8 bits (81), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 42/82 (51%), Gaps = 11/82 (13%)
Query 46 ARLQARTSLASLPAPRNDVTGQIPDDITE--EEKEL----LESEADLDMEEVERRRAQRA 99
+R R L LPAP+ND +P++ + EE+E+ +E AD+D + R A+R
Sbjct 472 SREHLRLGLLGLPAPKNDFEIVLPENAEKELEEREIDDTYIEDAADVDARKQAIRDAERV 531
Query 100 AEAARRRFLEQTQVMQRQLPRP 121
E R + +Q+ LPRP
Sbjct 532 KEMKR-----MHKAVQKDLPRP 548
> 7301749
Length=855
Score = 33.5 bits (75), Expect = 0.23, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 29/49 (59%), Gaps = 2/49 (4%)
Query 65 TGQIPDDITEEEKELLESEADLDMEEVERRRAQRAAEAARRRFLEQTQV 113
TG+ P+ + +E+++ EA + E E R + AEA +R FLE T++
Sbjct 82 TGESPE--APQPREMIDLEATFEKLENELREVNQNAEALKRNFLELTEL 128
> At3g07370
Length=278
Score = 31.2 bits (69), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 27/42 (64%), Gaps = 0/42 (0%)
Query 136 ASAIASAVQQQLQQDDDTIITEEGKDAAVAAISEAAALTENV 177
+ +ASA+ ++L++D + +E AA+ A +EA AL+ NV
Sbjct 2 VTGVASAMAERLKEDGNNCFKKERFGAAIDAYTEAIALSPNV 43
> At3g47920
Length=300
Score = 31.2 bits (69), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 26/49 (53%), Gaps = 0/49 (0%)
Query 93 RRRAQRAAEAARRRFLEQTQVMQRQLPRPILPPPKLLTSSIDAASAIAS 141
R RA E R Q+Q+ + P P+LPPP+++T ++ +A+
Sbjct 158 RERAVFHDERLRSSMNSQSQMTESSFPAPVLPPPRIVTPPLNHGEFLAA 206
> YMR213w
Length=590
Score = 30.4 bits (67), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 54 LASLPAPRNDVTGQIPDDITEEEKELLESEADLDME 89
ASLP+P+ND + +D EE+ E+ E E + + E
Sbjct 370 FASLPSPKNDFEIVLSEDEKEEDAEIAEYEKEFENE 405
> At5g17380
Length=572
Score = 29.6 bits (65), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 31/56 (55%), Gaps = 2/56 (3%)
Query 68 IPDDITEEEKELLESEADLDMEEVERRRAQRAAEAARRRFLEQTQVMQRQLPRPIL 123
IP D+ ++ + ESEAD ++EVER R + + R +E + R+ RP++
Sbjct 168 IPTDVLRQK--ISESEADKLVDEVERSRKEEPIRGSLRSEIESAVSLLRKAERPLI 221
> At4g08350
Length=1054
Score = 28.9 bits (63), Expect = 5.8, Method: Composition-based stats.
Identities = 19/64 (29%), Positives = 32/64 (50%), Gaps = 3/64 (4%)
Query 68 IPD---DITEEEKELLESEADLDMEEVERRRAQRAAEAARRRFLEQTQVMQRQLPRPILP 124
+PD D E + L E D D+E++ERR +R + + E+ +++Q P +
Sbjct 121 LPDERGDRRYERRFLPRDENDEDVEDLERRIQERFSSRHHEEYDEEATEVEQQALLPSVR 180
Query 125 PPKL 128
PKL
Sbjct 181 DPKL 184
> Hs18765739
Length=3657
Score = 28.5 bits (62), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 12/22 (54%), Positives = 16/22 (72%), Gaps = 0/22 (0%)
Query 57 LPAPRNDVTGQIPDDITEEEKE 78
LP +++V +PD ITEEEKE
Sbjct 1642 LPREKSEVQNLLPDTITEEEKE 1663
> Hs14719394
Length=3031
Score = 28.5 bits (62), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 12/22 (54%), Positives = 16/22 (72%), Gaps = 0/22 (0%)
Query 57 LPAPRNDVTGQIPDDITEEEKE 78
LP +++V +PD ITEEEKE
Sbjct 1016 LPREKSEVQNLLPDTITEEEKE 1037
> CE29213
Length=544
Score = 28.1 bits (61), Expect = 8.6, Method: Composition-based stats.
Identities = 24/95 (25%), Positives = 43/95 (45%), Gaps = 11/95 (11%)
Query 71 DITEEE--KELLESEADLDMEEV---ERRRAQRAAEAARRRFLEQTQVMQRQLPRPILPP 125
DI E+E +E+ + +A + E V ++R Q + R+R E + +RQ PP
Sbjct 29 DIGEDEDPEEVAQEDAGVWQERVPLTRQKRKQVLRKPGRKRTGEIIHIPKRQ------PP 82
Query 126 PKLLTSSIDAASAIASAVQQQLQQDDDTIITEEGK 160
+ S A V+Q L D ++ ++G+
Sbjct 83 RAVRVKSYQQYDAFDELVEQDLDDSDPEVMDDDGE 117
Lambda K H
0.307 0.122 0.318
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2779358582
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40