bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2255_orf1
Length=74
Score E
Sequences producing significant alignments: (Bits) Value
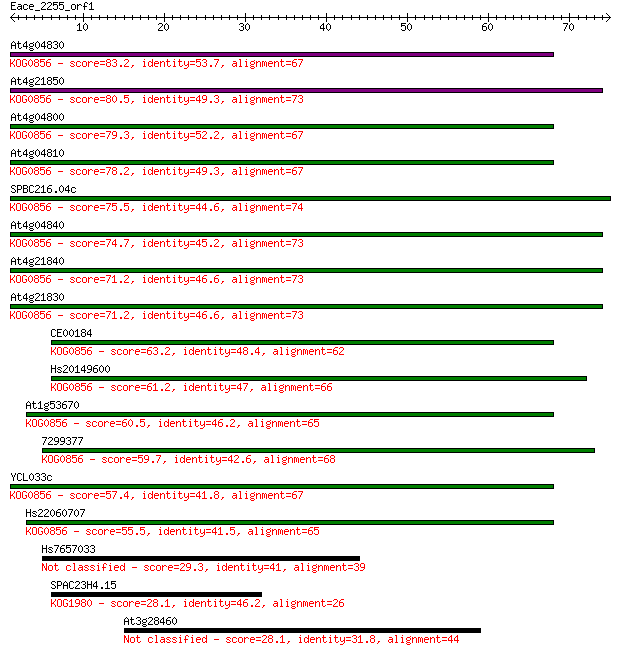
At4g04830 83.2 1e-16
At4g21850 80.5 7e-16
At4g04800 79.3 1e-15
At4g04810 78.2 4e-15
SPBC216.04c 75.5 2e-14
At4g04840 74.7 4e-14
At4g21840 71.2 4e-13
At4g21830 71.2 5e-13
CE00184 63.2 1e-10
Hs20149600 61.2 4e-10
At1g53670 60.5 8e-10
7299377 59.7 1e-09
YCL033c 57.4 6e-09
Hs22060707 55.5 3e-08
Hs7657033 29.3 2.1
SPAC23H4.15 28.1 4.1
At3g28460 28.1 4.4
> At4g04830
Length=139
Score = 83.2 bits (204), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 36/67 (53%), Positives = 51/67 (76%), Gaps = 1/67 (1%)
Query 1 QKTDEEWRQILTREEYRVLREKATEPPWSGEYNSVYPSSGFFVCRGCLSPLFPASSKFKS 60
QKT+EEWR +L+ E++R+LR+K TE P +GEY+ + G F C GC +PL+ +++KF S
Sbjct 9 QKTEEEWRAVLSPEQFRILRQKGTEKPGTGEYDKFF-EEGIFDCVGCKTPLYKSTTKFDS 67
Query 61 GCGWPAF 67
GCGWPAF
Sbjct 68 GCGWPAF 74
> At4g21850
Length=124
Score = 80.5 bits (197), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 36/73 (49%), Positives = 50/73 (68%), Gaps = 1/73 (1%)
Query 1 QKTDEEWRQILTREEYRVLREKATEPPWSGEYNSVYPSSGFFVCRGCLSPLFPASSKFKS 60
QK D++WR IL+ E++RVLREK TE GEY ++ G + C GC +PL+ +++KF S
Sbjct 16 QKKDQDWRAILSPEQFRVLREKGTENRGKGEYTKLF-DDGIYSCAGCATPLYKSTTKFDS 74
Query 61 GCGWPAFDRAYEG 73
GCGWP+F A G
Sbjct 75 GCGWPSFFDAIPG 87
> At4g04800
Length=136
Score = 79.3 bits (194), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 35/67 (52%), Positives = 50/67 (74%), Gaps = 1/67 (1%)
Query 1 QKTDEEWRQILTREEYRVLREKATEPPWSGEYNSVYPSSGFFVCRGCLSPLFPASSKFKS 60
QK DEEWR IL+ E++R+LR+K TE P +GEY + + G + C GC +PL+ +++KF +
Sbjct 8 QKGDEEWRAILSPEQFRILRQKGTEYPGTGEYVN-FDKEGVYGCVGCNAPLYKSTTKFNA 66
Query 61 GCGWPAF 67
GCGWPAF
Sbjct 67 GCGWPAF 73
> At4g04810
Length=139
Score = 78.2 bits (191), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 33/67 (49%), Positives = 50/67 (74%), Gaps = 1/67 (1%)
Query 1 QKTDEEWRQILTREEYRVLREKATEPPWSGEYNSVYPSSGFFVCRGCLSPLFPASSKFKS 60
+KT+EEWR +L+ E++R+LR+K TE P + EY+ + G F C GC +PL+ +++KF +
Sbjct 9 KKTEEEWRAVLSPEQFRILRQKGTETPGTEEYDKFF-EEGIFSCIGCKTPLYKSTTKFDA 67
Query 61 GCGWPAF 67
GCGWPAF
Sbjct 68 GCGWPAF 74
> SPBC216.04c
Length=138
Score = 75.5 bits (184), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 33/74 (44%), Positives = 49/74 (66%), Gaps = 0/74 (0%)
Query 1 QKTDEEWRQILTREEYRVLREKATEPPWSGEYNSVYPSSGFFVCRGCLSPLFPASSKFKS 60
+K+++EW++ L E+YR++R+K TE P +G + +P G FVC C L+ AS+KF+S
Sbjct 6 EKSEDEWKKELGPEKYRIMRQKGTEHPGAGRFTHQFPKQGVFVCAACKELLYKASTKFES 65
Query 61 GCGWPAFDRAYEGK 74
CGWPAF GK
Sbjct 66 HCGWPAFFDNLPGK 79
> At4g04840
Length=141
Score = 74.7 bits (182), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 33/73 (45%), Positives = 52/73 (71%), Gaps = 1/73 (1%)
Query 1 QKTDEEWRQILTREEYRVLREKATEPPWSGEYNSVYPSSGFFVCRGCLSPLFPASSKFKS 60
+K++EEWR +L+ E++++LREK+ E SGEY ++ G + C GC +P++ +++KF S
Sbjct 13 KKSNEEWRTVLSPEQFKILREKSIEKRGSGEYVKLF-EEGIYCCVGCGNPVYKSTTKFDS 71
Query 61 GCGWPAFDRAYEG 73
GCGWPAF A G
Sbjct 72 GCGWPAFFDAIPG 84
> At4g21840
Length=143
Score = 71.2 bits (173), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 34/73 (46%), Positives = 47/73 (64%), Gaps = 1/73 (1%)
Query 1 QKTDEEWRQILTREEYRVLREKATEPPWSGEYNSVYPSSGFFVCRGCLSPLFPASSKFKS 60
QK DEEWR +L+ E++RVLR K T+ GE+ + G + C GC + L+ +++KF S
Sbjct 15 QKQDEEWRAVLSPEQFRVLRLKGTDKRGKGEFTKKF-EEGTYSCAGCGTALYKSTTKFDS 73
Query 61 GCGWPAFDRAYEG 73
GCGWPAF A G
Sbjct 74 GCGWPAFFDAIPG 86
> At4g21830
Length=141
Score = 71.2 bits (173), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 34/73 (46%), Positives = 47/73 (64%), Gaps = 1/73 (1%)
Query 1 QKTDEEWRQILTREEYRVLREKATEPPWSGEYNSVYPSSGFFVCRGCLSPLFPASSKFKS 60
QK DEEWR +L+ E++RVLR K T+ GE+ + G + C GC + L+ +++KF S
Sbjct 13 QKQDEEWRAVLSPEQFRVLRLKGTDKRGKGEFTKKF-EEGTYSCAGCGTALYKSTTKFDS 71
Query 61 GCGWPAFDRAYEG 73
GCGWPAF A G
Sbjct 72 GCGWPAFFDAIPG 84
> CE00184
Length=152
Score = 63.2 bits (152), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 30/62 (48%), Positives = 39/62 (62%), Gaps = 1/62 (1%)
Query 6 EWRQILTREEYRVLREKATEPPWSGEYNSVYPSSGFFVCRGCLSPLFPASSKFKSGCGWP 65
EW+ +L E YRV RE TE P +G +N + G +VC C S LF + +KF +GCGWP
Sbjct 29 EWKSVLPNEVYRVARESGTETPHTGGFNDHF-EKGRYVCLCCGSELFNSDAKFWAGCGWP 87
Query 66 AF 67
AF
Sbjct 88 AF 89
> Hs20149600
Length=201
Score = 61.2 bits (147), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 43/66 (65%), Gaps = 1/66 (1%)
Query 6 EWRQILTREEYRVLREKATEPPWSGEYNSVYPSSGFFVCRGCLSPLFPASSKFKSGCGWP 65
EW++ LT E++ V REK TEPP+SG Y + +G + C C SPLF + K+ SG GWP
Sbjct 72 EWQKKLTPEQFYVTREKGTEPPFSGIYLN-NKEAGMYHCVCCDSPLFSSEKKYCSGTGWP 130
Query 66 AFDRAY 71
+F A+
Sbjct 131 SFSEAH 136
> At1g53670
Length=202
Score = 60.5 bits (145), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 30/66 (45%), Positives = 45/66 (68%), Gaps = 3/66 (4%)
Query 3 TDEEWRQILTREEYRVLREKATEPPWSGEY-NSVYPSSGFFVCRGCLSPLFPASSKFKSG 61
++ EW++ LT E+Y + R+K TE ++GEY NS P G + C C +PLF +S+KF SG
Sbjct 74 SENEWKKRLTPEQYYITRQKGTERAFTGEYWNSKTP--GVYNCVCCDTPLFDSSTKFDSG 131
Query 62 CGWPAF 67
GWP++
Sbjct 132 TGWPSY 137
> 7299377
Length=166
Score = 59.7 bits (143), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 29/68 (42%), Positives = 41/68 (60%), Gaps = 1/68 (1%)
Query 5 EEWRQILTREEYRVLREKATEPPWSGEYNSVYPSSGFFVCRGCLSPLFPASSKFKSGCGW 64
EE R+ LT +Y+V +E TE P++G YN Y G + C C LF + +K+ SGCGW
Sbjct 13 EELRKRLTPVQYQVTQEAGTERPFTGCYNKHY-EKGVYQCIVCHQDLFSSETKYDSGCGW 71
Query 65 PAFDRAYE 72
PAF+ +
Sbjct 72 PAFNDVLD 79
> YCL033c
Length=168
Score = 57.4 bits (137), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 28/67 (41%), Positives = 38/67 (56%), Gaps = 1/67 (1%)
Query 1 QKTDEEWRQILTREEYRVLREKATEPPWSGEYNSVYPSSGFFVCRGCLSPLFPASSKFKS 60
+ D +W LT + VLR+KATE P +G Y SG + C C PL+ + +KF +
Sbjct 37 ESNDVKWNDALTPLQLMVLRDKATERPNTGAYLHT-NESGVYHCANCDRPLYSSKAKFDA 95
Query 61 GCGWPAF 67
CGWPAF
Sbjct 96 RCGWPAF 102
> Hs22060707
Length=185
Score = 55.5 bits (132), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 27/65 (41%), Positives = 39/65 (60%), Gaps = 1/65 (1%)
Query 3 TDEEWRQILTREEYRVLREKATEPPWSGEYNSVYPSSGFFVCRGCLSPLFPASSKFKSGC 62
+ +E R+ LT +Y V +EK TE + GEY + G + C C +PLF + +KF SG
Sbjct 39 SQQELRKRLTPLQYHVTQEKGTESAFEGEYTH-HKDPGIYKCVVCGTPLFKSETKFDSGS 97
Query 63 GWPAF 67
GWP+F
Sbjct 98 GWPSF 102
> Hs7657033
Length=201
Score = 29.3 bits (64), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 5/39 (12%)
Query 5 EEWRQILTREEYRVLREKATEPPWSGEYNSVYPSSGFFV 43
E+ R L RE+YR LR P + + SVY + GFF+
Sbjct 39 EQRRGFLAREQYRALR-----PDLADKVASVYEAPGFFL 72
> SPAC23H4.15
Length=783
Score = 28.1 bits (61), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 6 EWRQILTREEYRVLREKATEPPWSGE 31
EWRQ+ E YR L+ K + P+ GE
Sbjct 516 EWRQLFKFENYRNLKNKFLKQPFIGE 541
> At3g28460
Length=307
Score = 28.1 bits (61), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 24/45 (53%), Gaps = 1/45 (2%)
Query 15 EYRVLREKATEPPWSGEYNSVYPS-SGFFVCRGCLSPLFPASSKF 58
+Y VL ++ + P GE +YPS + GCL ++P +S +
Sbjct 247 DYEVLMDQIAKSPAIGENTFIYPSRTTMLDSCGCLEKVWPDTSSY 291
Lambda K H
0.317 0.135 0.453
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1184572648
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40