bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2237_orf1
Length=77
Score E
Sequences producing significant alignments: (Bits) Value
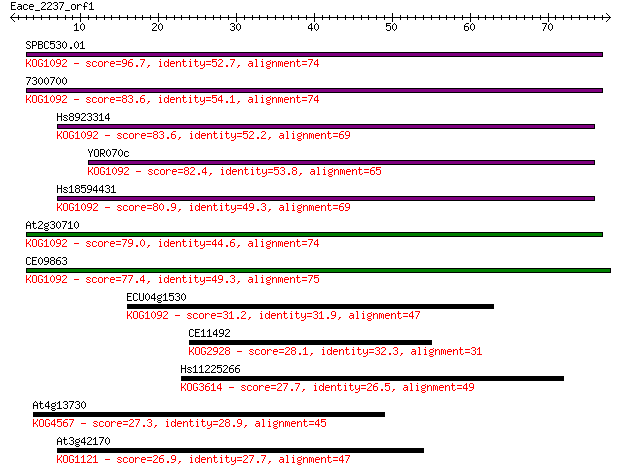
SPBC530.01 96.7 9e-21
7300700 83.6 8e-17
Hs8923314 83.6 9e-17
YOR070c 82.4 2e-16
Hs18594431 80.9 6e-16
At2g30710 79.0 2e-15
CE09863 77.4 5e-15
ECU04g1530 31.2 0.44
CE11492 28.1 4.3
Hs11225266 27.7 5.3
At4g13730 27.3 7.7
At3g42170 26.9 8.2
> SPBC530.01
Length=514
Score = 96.7 bits (239), Expect = 9e-21, Method: Composition-based stats.
Identities = 39/74 (52%), Positives = 57/74 (77%), Gaps = 0/74 (0%)
Query 3 YLCEQAEGFSDFHVYVCAVFLVNWSRQLKQMDFQQMIMFIQNFPTTTWGSQEIETLLAEA 62
Y+ E +GFS+FH+YVCA FLV WS +L++M+FQ +++F+Q+ PT W +++IE LL+EA
Sbjct 439 YMAEGVQGFSEFHLYVCAAFLVKWSSELQKMEFQDILIFLQSIPTKDWSTKDIEILLSEA 498
Query 63 FVLKSLYHSAPKHL 76
F+ KSLY A HL
Sbjct 499 FLWKSLYSGAGAHL 512
> 7300700
Length=545
Score = 83.6 bits (205), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 40/75 (53%), Positives = 53/75 (70%), Gaps = 2/75 (2%)
Query 3 YLCEQAEGFSDFHVYVCAVFLVNWSRQL-KQMDFQQMIMFIQNFPTTTWGSQEIETLLAE 61
YL E ++GF+ FH+YVCA FL++W QL +Q DFQ +++ +QN PT W ++I LLAE
Sbjct 468 YLAE-SDGFALFHLYVCAAFLLHWKEQLMQQNDFQGLMLLLQNLPTHNWSDRQINVLLAE 526
Query 62 AFVLKSLYHSAPKHL 76
AF LK Y APKHL
Sbjct 527 AFRLKFTYADAPKHL 541
> Hs8923314
Length=163
Score = 83.6 bits (205), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 36/70 (51%), Positives = 50/70 (71%), Gaps = 1/70 (1%)
Query 7 QAEGFSDFHVYVCAVFLVNWSRQ-LKQMDFQQMIMFIQNFPTTTWGSQEIETLLAEAFVL 65
+ EGFS FH+YVCA FL+ W ++ L + DFQ ++M +QN PT WG++EI LLAEA+ L
Sbjct 91 EPEGFSHFHLYVCAAFLIKWRKEILDEEDFQGLLMLLQNLPTIHWGNEEIGLLLAEAYRL 150
Query 66 KSLYHSAPKH 75
K ++ AP H
Sbjct 151 KYMFADAPNH 160
> YOR070c
Length=637
Score = 82.4 bits (202), Expect = 2e-16, Method: Composition-based stats.
Identities = 35/65 (53%), Positives = 46/65 (70%), Gaps = 0/65 (0%)
Query 11 FSDFHVYVCAVFLVNWSRQLKQMDFQQMIMFIQNFPTTTWGSQEIETLLAEAFVLKSLYH 70
++FHV+VCA FL+ WS QL +MDFQ+ I F+QN PT W +IE LL+EAF+ +SLY
Sbjct 571 LNEFHVFVCAAFLIKWSDQLMEMDFQETITFLQNPPTKDWTETDIEMLLSEAFIWQSLYK 630
Query 71 SAPKH 75
A H
Sbjct 631 DATSH 635
> Hs18594431
Length=517
Score = 80.9 bits (198), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 34/70 (48%), Positives = 50/70 (71%), Gaps = 1/70 (1%)
Query 7 QAEGFSDFHVYVCAVFLVNWSRQ-LKQMDFQQMIMFIQNFPTTTWGSQEIETLLAEAFVL 65
+ +GFS FH+YVCA FLV W ++ L++ DFQ++++F+QN PT W ++I LLAEA+ L
Sbjct 445 EPDGFSHFHLYVCAAFLVRWRKEILEEKDFQELLLFLQNLPTAHWDDEDISLLLAEAYRL 504
Query 66 KSLYHSAPKH 75
K + AP H
Sbjct 505 KFAFADAPNH 514
> At2g30710
Length=343
Score = 79.0 bits (193), Expect = 2e-15, Method: Composition-based stats.
Identities = 33/74 (44%), Positives = 53/74 (71%), Gaps = 1/74 (1%)
Query 3 YLCEQAEGFSDFHVYVCAVFLVNWSRQLKQMDFQQMIMFIQNFPTTTWGSQEIETLLAEA 62
YL E + DF VY+ A FL+ WS +LK++DFQ+M+MF+Q+ PT W QE+E +L+ A
Sbjct 269 YLAE-GDALPDFLVYIYASFLLTWSDELKKLDFQEMVMFLQHLPTHNWSDQELEMVLSRA 327
Query 63 FVLKSLYHSAPKHL 76
++ S+++++P HL
Sbjct 328 YMWHSMFNNSPNHL 341
> CE09863
Length=495
Score = 77.4 bits (189), Expect = 5e-15, Method: Composition-based stats.
Identities = 37/76 (48%), Positives = 53/76 (69%), Gaps = 2/76 (2%)
Query 3 YLCEQAEGFSDFHVYVCAVFLVNWSRQLK-QMDFQQMIMFIQNFPTTTWGSQEIETLLAE 61
YL E +GF FH YVCA FL WS+QL+ + DFQ +++ +QN PT +WG +EI L A+
Sbjct 415 YLSE-PDGFMQFHNYVCAAFLRTWSKQLQAEKDFQGVMILLQNLPTQSWGDREICELTAD 473
Query 62 AFVLKSLYHSAPKHLN 77
AF L+S++ A +HL+
Sbjct 474 AFSLQSVFDGARRHLS 489
> ECU04g1530
Length=329
Score = 31.2 bits (69), Expect = 0.44, Method: Composition-based stats.
Identities = 15/47 (31%), Positives = 23/47 (48%), Gaps = 0/47 (0%)
Query 16 VYVCAVFLVNWSRQLKQMDFQQMIMFIQNFPTTTWGSQEIETLLAEA 62
VY L+ + L + DF I+F+Q+ W EIE +L+ A
Sbjct 265 VYFGVALLMKFKSTLVENDFSHNILFLQSIYDREWEEAEIELILSSA 311
> CE11492
Length=430
Score = 28.1 bits (61), Expect = 4.3, Method: Composition-based stats.
Identities = 10/32 (31%), Positives = 20/32 (62%), Gaps = 1/32 (3%)
Query 24 VNWSRQLK-QMDFQQMIMFIQNFPTTTWGSQE 54
++W+R ++ +M+ QQ+I+ I N W S +
Sbjct 221 ISWARSIRRKMNSQQLILIIDNIEAPDWRSDQ 252
> Hs11225266
Length=1165
Score = 27.7 bits (60), Expect = 5.3, Method: Composition-based stats.
Identities = 13/49 (26%), Positives = 28/49 (57%), Gaps = 2/49 (4%)
Query 23 LVNWSRQLKQMDFQQMIMFIQNFPTTTWGSQEIETLLAEAFVLKSLYHS 71
+V W++ L+ + Q ++ + +F GS+E++T++ +A V HS
Sbjct 296 IVRWTKLLQNITSHQHLLTVYDFEQE--GSEELDTVILKALVKACKSHS 342
> At4g13730
Length=449
Score = 27.3 bits (59), Expect = 7.7, Method: Composition-based stats.
Identities = 13/45 (28%), Positives = 21/45 (46%), Gaps = 0/45 (0%)
Query 4 LCEQAEGFSDFHVYVCAVFLVNWSRQLKQMDFQQMIMFIQNFPTT 48
L EG + + +C L+ R+L DF + +QN+P T
Sbjct 390 LLSDPEGPQETLLRICCAMLILVRRRLLAGDFTSNLKLLQNYPPT 434
> At3g42170
Length=696
Score = 26.9 bits (58), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 13/47 (27%), Positives = 21/47 (44%), Gaps = 0/47 (0%)
Query 7 QAEGFSDFHVYVCAVFLVNWSRQLKQMDFQQMIMFIQNFPTTTWGSQ 53
+A+G SDF Y+ N +L Q + ++ +Q F W Q
Sbjct 570 KADGLSDFDTYIMETTGQNLKSELDQYLDETLLPRVQEFDVLDWWKQ 616
Lambda K H
0.326 0.134 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1175087368
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40