bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2220_orf2
Length=111
Score E
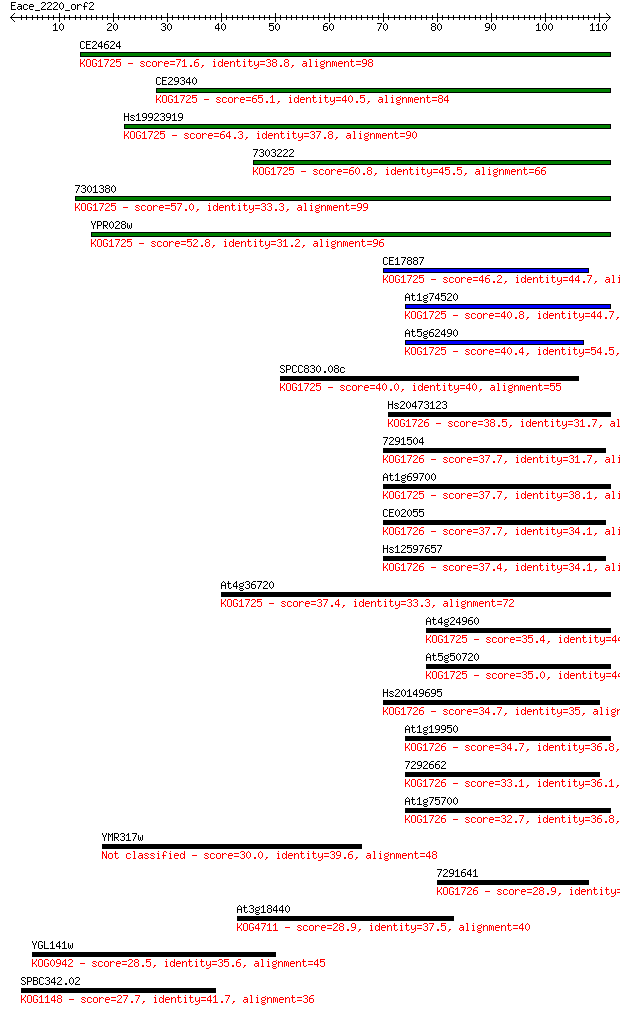
Sequences producing significant alignments: (Bits) Value
CE24624 71.6 3e-13
CE29340 65.1 3e-11
Hs19923919 64.3 5e-11
7303222 60.8 6e-10
7301380 57.0 8e-09
YPR028w 52.8 2e-07
CE17887 46.2 2e-05
At1g74520 40.8 6e-04
At5g62490 40.4 7e-04
SPCC830.08c 40.0 0.001
Hs20473123 38.5 0.003
7291504 37.7 0.005
At1g69700 37.7 0.006
CE02055 37.7 0.006
Hs12597657 37.4 0.006
At4g36720 37.4 0.007
At4g24960 35.4 0.024
At5g50720 35.0 0.033
Hs20149695 34.7 0.043
At1g19950 34.7 0.045
7292662 33.1 0.15
At1g75700 32.7 0.19
YMR317w 30.0 1.1
7291641 28.9 2.4
At3g18440 28.9 2.7
YGL141w 28.5 2.8
SPBC342.02 27.7 5.1
> CE24624
Length=183
Score = 71.6 bits (174), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 38/106 (35%), Positives = 59/106 (55%), Gaps = 12/106 (11%)
Query 14 PAAAAVRRLFSNLDKYLED--------ATLAQQASSFTGLRPSVLVIALGLFLIISLGTG 65
P V+++ ++DK L + AT+ Q+ TG++ LV+ + + L G
Sbjct 2 PVPPQVQKVLDDVDKQLHEPSTVTNVLATVEQK----TGVKRLHLVLGVVGLQALYLIFG 57
Query 66 IGGGLVCDVAGFLYPAWMSFRAIESAGAEDDKLWLTYWVVYGAFSI 111
LVC+ GF+YPA+MS +AIES+ EDD WLTYWV++ S+
Sbjct 58 HSAQLVCNFMGFVYPAYMSIKAIESSNKEDDTQWLTYWVIFAILSV 103
> CE29340
Length=207
Score = 65.1 bits (157), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 34/84 (40%), Positives = 47/84 (55%), Gaps = 9/84 (10%)
Query 28 KYLEDATLAQQASSFTGLRPSVLVIALGLFLIISLGTGIGGGLVCDVAGFLYPAWMSFRA 87
K LEDAT GL+ +L L + + G G VC++ G YPA++S +A
Sbjct 46 KKLEDAT---------GLKREMLAYGLIGLNCVYMIIGSGAEFVCNLIGVAYPAYVSVKA 96
Query 88 IESAGAEDDKLWLTYWVVYGAFSI 111
I + G +DD +WL YW V+GAFSI
Sbjct 97 IRTEGTDDDTMWLIYWTVFGAFSI 120
> Hs19923919
Length=184
Score = 64.3 bits (155), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 34/94 (36%), Positives = 50/94 (53%), Gaps = 4/94 (4%)
Query 22 LFSNLDKYLEDATLAQQA----SSFTGLRPSVLVIALGLFLIISLGTGIGGGLVCDVAGF 77
L ++ +LE L + + TG+ L L + L G G L+C++ GF
Sbjct 4 LRQRVEHFLEQRNLVTEVLGALEAKTGVEKRYLAAGAVTLLSLYLLFGYGASLLCNLIGF 63
Query 78 LYPAWMSFRAIESAGAEDDKLWLTYWVVYGAFSI 111
+YPA+ S +AIES +DD +WLTYWVVY F +
Sbjct 64 VYPAYASIKAIESPSKDDDTVWLTYWVVYALFGL 97
> 7303222
Length=178
Score = 60.8 bits (146), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 30/66 (45%), Positives = 43/66 (65%), Gaps = 1/66 (1%)
Query 46 RPSVLVIALGLFLIISLGTGIGGGLVCDVAGFLYPAWMSFRAIESAGAEDDKLWLTYWVV 105
R ++ V A+GL I L G G L+C++ G LYPA++S AIES+ +DD WL YWV
Sbjct 42 RVNIFVGAVGL-CAIYLIFGWGAQLLCNIIGVLYPAYISIHAIESSTKQDDTKWLIYWVT 100
Query 106 YGAFSI 111
+G F++
Sbjct 101 FGIFTV 106
> 7301380
Length=174
Score = 57.0 bits (136), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 33/108 (30%), Positives = 54/108 (50%), Gaps = 9/108 (8%)
Query 13 APAAAAVRRLFS----NLDKYLEDATLA-----QQASSFTGLRPSVLVIALGLFLIISLG 63
A + V+R F+ ++++ L DA+ TG+ + + LF L
Sbjct 2 AAQVSQVQRFFNGYKQDINRSLRDASKPWTKAFDSLEEKTGVERVNIFVGSILFCAFYLV 61
Query 64 TGIGGGLVCDVAGFLYPAWMSFRAIESAGAEDDKLWLTYWVVYGAFSI 111
G L+C++ G LYPA++S IES+ +DD WL YWV +G F++
Sbjct 62 FGWCAQLLCNIIGVLYPAYISIHGIESSTKQDDIRWLIYWVTFGIFTV 109
> YPR028w
Length=180
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/98 (30%), Positives = 50/98 (51%), Gaps = 2/98 (2%)
Query 16 AAAVRRLFSNLDKYLEDATLAQQASSFTGLRPSVLVIALGL--FLIISLGTGIGGGLVCD 73
A+++ D + QQ + T L S LV LG L+I + G G ++ +
Sbjct 5 ASSIHSQMKQFDTKYSGNRILQQLENKTNLPKSYLVAGLGFAYLLLIFINVGGVGEILSN 64
Query 74 VAGFLYPAWMSFRAIESAGAEDDKLWLTYWVVYGAFSI 111
AGF+ PA++S A+++ + DD LTYW+V+ S+
Sbjct 65 FAGFVLPAYLSLVALKTPTSTDDTQLLTYWIVFSFLSV 102
> CE17887
Length=205
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 25/38 (65%), Gaps = 0/38 (0%)
Query 70 LVCDVAGFLYPAWMSFRAIESAGAEDDKLWLTYWVVYG 107
L+C++ GF YP + S +AI S G +DD +WL YW +
Sbjct 102 LLCNLIGFGYPTYASVKAIRSPGGDDDTVWLIYWTCFA 139
> At1g74520
Length=177
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 74 VAGFLYPAWMSFRAIESAGAEDDKLWLTYWVVYGAFSI 111
V +YP + S +AIE+ DDK WLTYWV+Y ++
Sbjct 23 VVSLVYPLYASVQAIETQSHADDKQWLTYWVLYSLLTL 60
> At5g62490
Length=167
Score = 40.4 bits (93), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 18/33 (54%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 74 VAGFLYPAWMSFRAIESAGAEDDKLWLTYWVVY 106
V +YP + S RAIES DDK WLTYW +Y
Sbjct 23 VISLVYPLYASVRAIESRSHGDDKQWLTYWALY 55
> SPCC830.08c
Length=182
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/55 (40%), Positives = 30/55 (54%), Gaps = 6/55 (10%)
Query 51 VIALGLFLIISLGTGIGGGLVCDVAGFLYPAWMSFRAIESAGAEDDKLWLTYWVV 105
+ AL LFL GG L+ ++ F PA+ S AIE+ DD WLTY++V
Sbjct 45 IYALFLFL------NWGGFLLTNLLAFAMPAFFSINAIETTNKADDTQWLTYYLV 93
> Hs20473123
Length=255
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 13/41 (31%), Positives = 27/41 (65%), Gaps = 0/41 (0%)
Query 71 VCDVAGFLYPAWMSFRAIESAGAEDDKLWLTYWVVYGAFSI 111
V V G LYPA+ S++A+++ ++ W+ YW+V+ +++
Sbjct 10 VVLVFGMLYPAYYSYKAVKTKNVKEYVRWMMYWIVFALYTV 50
> 7291504
Length=569
Score = 37.7 bits (86), Expect = 0.005, Method: Composition-based stats.
Identities = 13/41 (31%), Positives = 25/41 (60%), Gaps = 0/41 (0%)
Query 70 LVCDVAGFLYPAWMSFRAIESAGAEDDKLWLTYWVVYGAFS 110
L+ G LYPA+ S++A+ + ++ W+ YW+V+ F+
Sbjct 9 LIILFCGTLYPAYASYKAVRTKDVKEYVKWMMYWIVFAFFT 49
> At1g69700
Length=184
Score = 37.7 bits (86), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 25/42 (59%), Gaps = 0/42 (0%)
Query 70 LVCDVAGFLYPAWMSFRAIESAGAEDDKLWLTYWVVYGAFSI 111
L + +YP + S +AIE+ +D+ WLTYWV+Y S+
Sbjct 21 LALPLVTLVYPLYASVKAIETRSLPEDEQWLTYWVLYALISL 62
> CE02055
Length=229
Score = 37.7 bits (86), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 25/41 (60%), Gaps = 0/41 (0%)
Query 70 LVCDVAGFLYPAWMSFRAIESAGAEDDKLWLTYWVVYGAFS 110
L+ AG LYPA+ S++A+ + + W+ YW+V+ +S
Sbjct 8 LLIITAGTLYPAYRSYKAVRTKDTREYVKWMMYWIVFAIYS 48
> Hs12597657
Length=201
Score = 37.4 bits (85), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 27/41 (65%), Gaps = 0/41 (0%)
Query 70 LVCDVAGFLYPAWMSFRAIESAGAEDDKLWLTYWVVYGAFS 110
LV + G LYPA+ S++A++S ++ W+ YW+++ F+
Sbjct 9 LVVLIFGTLYPAYYSYKAVKSKDIKEYVKWMMYWIIFALFT 49
> At4g36720
Length=227
Score = 37.4 bits (85), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 35/79 (44%), Gaps = 7/79 (8%)
Query 40 SSFTGLRPSVLVIALGL-------FLIISLGTGIGGGLVCDVAGFLYPAWMSFRAIESAG 92
S F PS+ ++ GL L L + I C G P + +F+AIES
Sbjct 2 SGFASQIPSMALLGSGLTGEVGLRVLFSPLSSNIVLRTACCSIGIGLPVYSTFKAIESGD 61
Query 93 AEDDKLWLTYWVVYGAFSI 111
+ + L YW YG+FS+
Sbjct 62 ENEQQKMLIYWAAYGSFSL 80
> At4g24960
Length=116
Score = 35.4 bits (80), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 78 LYPAWMSFRAIESAGAEDDKLWLTYWVVYGAFSI 111
LYP + S A+ES DD+ WL YW++Y S+
Sbjct 3 LYPLYASVIAMESTTKVDDEQWLAYWIIYSFLSL 36
> At5g50720
Length=107
Score = 35.0 bits (79), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 78 LYPAWMSFRAIESAGAEDDKLWLTYWVVYGAFSI 111
LYP + S AIES DD+ WL YW++Y ++
Sbjct 3 LYPLYASVIAIESPSKVDDEQWLAYWILYSFLTL 36
> Hs20149695
Length=257
Score = 34.7 bits (78), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 24/40 (60%), Gaps = 0/40 (0%)
Query 70 LVCDVAGFLYPAWMSFRAIESAGAEDDKLWLTYWVVYGAF 109
LV V G L PA+ S++A+++ + W+ YW+V+ F
Sbjct 9 LVVLVFGMLCPAYASYKAVKTKNIREYVRWMMYWIVFALF 48
> At1g19950
Length=304
Score = 34.7 bits (78), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 23/40 (57%), Gaps = 2/40 (5%)
Query 74 VAGFLYPAWMSFRAIESAGAEDDKL--WLTYWVVYGAFSI 111
V G+ YPA+ ++A+E E +L W YW++ A +I
Sbjct 2 VFGYAYPAYECYKAVEKNKPEMQQLRFWCQYWILVAALTI 41
> 7292662
Length=240
Score = 33.1 bits (74), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 22/38 (57%), Gaps = 2/38 (5%)
Query 74 VAGFLYPAWMSFRAIESAGAEDDKL--WLTYWVVYGAF 109
+ G YPA+ S++ ++S + L WL YW+ YG +
Sbjct 13 LVGCFYPAFASYKILKSQNCSVNDLRGWLIYWIAYGVY 50
> At1g75700
Length=166
Score = 32.7 bits (73), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 22/40 (55%), Gaps = 2/40 (5%)
Query 74 VAGFLYPAWMSFRAIESAGAEDDKL--WLTYWVVYGAFSI 111
V G+ YPA+ F+ +E E +L W YW++ A +I
Sbjct 2 VFGYAYPAYECFKTVELNKPEIQQLQFWCQYWIIVAALTI 41
> YMR317w
Length=1140
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 26/49 (53%), Gaps = 1/49 (2%)
Query 18 AVRRLFSNLDKYLEDATLAQQASSFTG-LRPSVLVIALGLFLIISLGTG 65
A RLFS+ + + +A +ASS T LRPS +A + SL TG
Sbjct 452 ASSRLFSSKNTSVTSTLVATEASSVTSSLRPSSETLASNSIIESSLSTG 500
> 7291641
Length=279
Score = 28.9 bits (63), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 11/28 (39%), Positives = 17/28 (60%), Gaps = 1/28 (3%)
Query 80 PAWMSFRAIESAGAEDDKLWLTYWVVYG 107
PAW +++ + S G E+ W YW+VY
Sbjct 3 PAWQTYKTLNS-GDEEFLAWAKYWIVYA 29
> At3g18440
Length=598
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 15/41 (36%), Positives = 23/41 (56%), Gaps = 5/41 (12%)
Query 43 TGLRPSVLV-IALGLFLIISLGTGIGGGLVCDVAGFLYPAW 82
+G R + +A+ FL+I+LG G+ G V F+YP W
Sbjct 212 SGFRTGQFIEVAISRFLLIALGAGVSLG----VNMFIYPIW 248
> YGL141w
Length=910
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 16/45 (35%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 5 VSHSLSSSAPAAAAVRRLFSNLDKYLEDATLAQQASSFTGLRPSV 49
+SH+ SS P + R+L S L Y +DA +A+ S P+V
Sbjct 99 LSHNFLSSYPNSLGNRQLLSLLKLYQDDALVAETLSDLNMDCPTV 143
> SPBC342.02
Length=811
Score = 27.7 bits (60), Expect = 5.1, Method: Composition-based stats.
Identities = 15/36 (41%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 3 QYVSHSLSSSAPAAAAVRRLFSNLDKYLEDATLAQQ 38
Q+VS S++P A RLF+NL K A L +Q
Sbjct 687 QWVSRDKESNSPVLIAETRLFNNLFKCDNPAALKEQ 722
Lambda K H
0.322 0.135 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1192473466
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40