bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2208_orf1
Length=126
Score E
Sequences producing significant alignments: (Bits) Value
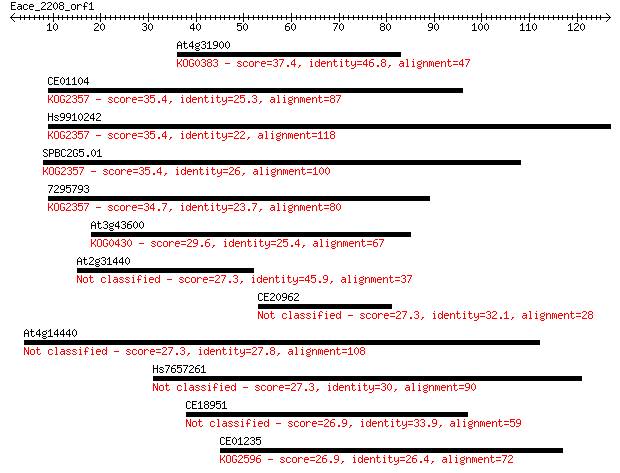
At4g31900 37.4 0.006
CE01104 35.4 0.025
Hs9910242 35.4 0.025
SPBC2G5.01 35.4 0.026
7295793 34.7 0.039
At3g43600 29.6 1.5
At2g31440 27.3 6.4
CE20962 27.3 7.2
At4g14440 27.3 7.5
Hs7657261 27.3 7.5
CE18951 26.9 9.6
CE01235 26.9 9.6
> At4g31900
Length=1067
Score = 37.4 bits (85), Expect = 0.006, Method: Composition-based stats.
Identities = 22/47 (46%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 36 KLRSSSSSSNLLPSGLVLFSDTNEAAEKLLETPGVLKLLQQLAPHLK 82
KLR S LLP F D NEA KLLE G L+LL ++ LK
Sbjct 381 KLRQVCSHPYLLPDFEPRFEDANEAFTKLLEASGKLQLLDKMMVKLK 427
> CE01104
Length=442
Score = 35.4 bits (80), Expect = 0.025, Method: Composition-based stats.
Identities = 22/87 (25%), Positives = 41/87 (47%), Gaps = 1/87 (1%)
Query 9 EAFIVAVCRKEEQRNTLENHWDIAEFCKLRSSSSSSNLLPSGLVLFSDTNEAAEKLLETP 68
+ I AV K+ + D+ F R ++ LP+ +++D NE +L+ P
Sbjct 226 DPLIFAVGEKKIASKYFKEMLDLNSFASERKQAAQQFNLPASWQVYADQNEVVFSILD-P 284
Query 69 GVLKLLQQLAPHLKCMYTSDLCTNTNP 95
GV+ LL++ ++ ++ SD T P
Sbjct 285 GVVSLLKKHEDAIEFIHISDQFTGPKP 311
> Hs9910242
Length=483
Score = 35.4 bits (80), Expect = 0.025, Method: Composition-based stats.
Identities = 26/118 (22%), Positives = 52/118 (44%), Gaps = 8/118 (6%)
Query 9 EAFIVAVCRKEEQRNTLENHWDIAEFCKLRSSSSSSNLLPSGLVLFSDTNEAAEKLLETP 68
+ ++ AV ++ + D++EFC + S + LP L + S+ E + +++T
Sbjct 257 DTYVFAVGTRKALVRLQKEMQDLSEFCSDKPKSGAKYGLPDSLAILSEMGEVTDGMMDTK 316
Query 69 GVLKLLQQLAPHLKCMYTSDLCTNTNPTLAPTIAALYGTAAAAPNKPKRVLRLTYYLP 126
++ L A ++ ++ SD + P I G P+ KR L T+ +P
Sbjct 317 -MVHFLTHYADKIESVHFSDQFS------GPKIMQEEGQPLKLPDT-KRTLLFTFNVP 366
> SPBC2G5.01
Length=374
Score = 35.4 bits (80), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 26/100 (26%), Positives = 46/100 (46%), Gaps = 10/100 (10%)
Query 8 GEAFIVAVCRKEEQRNTLENHWDIAEFCKLRSSSSSSNLLPSGLVLFSDTNEAAEKLLET 67
E F+ A+ K+ R E +D++ F ++ SSS LP VL S+ NE ++ + E
Sbjct 177 AERFVFAIVHKDCMRILREIRYDLS-FTRI----SSSPYLPETHVLMSENNECSQAIFEI 231
Query 68 PGVLKLLQQLAPHLKCMYTSDLCTNTNPTLAPTIAALYGT 107
P + + + +L+ +D P++ P Y T
Sbjct 232 PEFMSSINECIENLEYFIVTD-----QPSVPPATEKDYVT 266
> 7295793
Length=476
Score = 34.7 bits (78), Expect = 0.039, Method: Composition-based stats.
Identities = 19/80 (23%), Positives = 40/80 (50%), Gaps = 1/80 (1%)
Query 9 EAFIVAVCRKEEQRNTLENHWDIAEFCKLRSSSSSSNLLPSGLVLFSDTNEAAEKLLETP 68
+AF+ AV K+ + + D+++FC L +P+G + S+ EA +LE+
Sbjct 259 DAFVFAVGTKKTITKLFKEYTDLSKFCSLVGKPEDRYNVPTGFGVLSEIPEATSAILESR 318
Query 69 GVLKLLQQLAPHLKCMYTSD 88
++ L + ++ ++ SD
Sbjct 319 -IITALNKYQSYIDYLHISD 337
> At3g43600
Length=1321
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 17/68 (25%), Positives = 34/68 (50%), Gaps = 1/68 (1%)
Query 18 KEEQRNTLENHWDIAEFCKLRSSSSSSNLLPSG-LVLFSDTNEAAEKLLETPGVLKLLQQ 76
K+E+ + + DI L+ + N + G +V S A +++ +PGV K+ +
Sbjct 260 KDEREQNYDKYIDITRIPHLKEIRENQNGVEIGSVVTISKVIAALKEIRVSPGVEKIFGK 319
Query 77 LAPHLKCM 84
LA H++ +
Sbjct 320 LATHMEMI 327
> At2g31440
Length=127
Score = 27.3 bits (59), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 23/44 (52%), Gaps = 7/44 (15%)
Query 15 VCRKEEQR-NTLENH----W--DIAEFCKLRSSSSSSNLLPSGL 51
VC++E+ R N +H W DI EFC R S+ LP G+
Sbjct 25 VCKREQSRSNNSSSHTPYCWNVDIGEFCIGRLCDRGSSSLPCGI 68
> CE20962
Length=326
Score = 27.3 bits (59), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 9/28 (32%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 53 LFSDTNEAAEKLLETPGVLKLLQQLAPH 80
+F+D +AE+ + ++K++ QL+PH
Sbjct 129 VFTDLRFSAERAINITSIIKVVDQLSPH 156
> At4g14440
Length=238
Score = 27.3 bits (59), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 30/118 (25%), Positives = 49/118 (41%), Gaps = 22/118 (18%)
Query 4 LPPAGEAFIVAVCRKEEQR---NTLENHWDIAEFCKLRSSSSSSNLLPSGLVLFSDTNEA 60
L G+ F++ + ++E R +T+ + + E K +S+ S + FS+ +
Sbjct 4 LEKRGDLFLLTLTGEDEHRFHPDTIASVLSLLEQAKSQSTKGSVLITTGHGKFFSNGFDL 63
Query 61 AEKLLETPGVLKLLQQLAPHLKCMYTSDLCTNTNPTLA-------PTIAALYGTAAAA 111
A G +K + Q+ K P LA PTIAAL G AAA+
Sbjct 64 AWAQSAGHGAIKRMHQMVKSFK------------PVLAALLDLPMPTIAALNGHAAAS 109
> Hs7657261
Length=1499
Score = 27.3 bits (59), Expect = 7.5, Method: Composition-based stats.
Identities = 27/98 (27%), Positives = 43/98 (43%), Gaps = 9/98 (9%)
Query 31 IAEFCKLRSSSSSSNLLPSGLVLFSDTNEAAEKLLETPGVLKLLQQLAPHLKCMYTSDLC 90
I + KLRS +S +NL V+ +T A ++L LK ++ + +K C
Sbjct 742 IDDLFKLRSKTSCANLKRFEEVINQETFWVASEILRETNQLKRMKIIKHFIKIALHCREC 801
Query 91 TNTNP--------TLAPTIAALYGTAAAAPNKPKRVLR 120
N N LAP +A L T PNK +++ +
Sbjct 802 KNFNSMFAIISGLNLAP-VARLRTTWEKLPNKYEKLFQ 838
> CE18951
Length=821
Score = 26.9 bits (58), Expect = 9.6, Method: Composition-based stats.
Identities = 20/68 (29%), Positives = 33/68 (48%), Gaps = 9/68 (13%)
Query 38 RSSSSSSNLLPSGLVLFSDTNEAAEKLLETP--------GVLKLL-QQLAPHLKCMYTSD 88
RSS S++ + L S ++ K TP GVL +++ PHLKC Y S+
Sbjct 727 RSSLQRSSMRSESIQLASPSSAGEFKQPFTPSGVTKERVGVLTARNEKVKPHLKCSYASE 786
Query 89 LCTNTNPT 96
+ + +P+
Sbjct 787 VGSTNSPS 794
> CE01235
Length=470
Score = 26.9 bits (58), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 32/72 (44%), Gaps = 1/72 (1%)
Query 45 NLLPSGLVLFSDTNEAAEKLLETPGVLKLLQQLAPHLKCMYTSDLCTNTNPTLAPTIAAL 104
+L +GLV+ + + KL++ + + LA HL+ T+ NT L P +
Sbjct 131 DLSVAGLVIVKNGEKLQHKLIDVKKPVLFIPNLAIHLETDRTT-FKPNTETELRPILETF 189
Query 105 YGTAAAAPNKPK 116
AP KP+
Sbjct 190 AAAGINAPQKPE 201
Lambda K H
0.317 0.131 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1180352192
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40