bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2196_orf1
Length=102
Score E
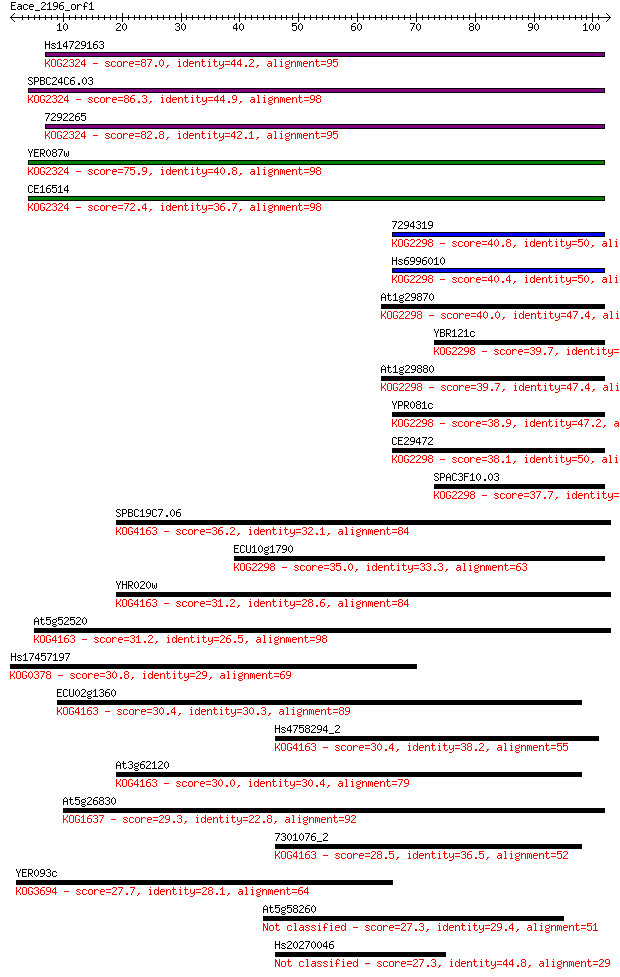
Sequences producing significant alignments: (Bits) Value
Hs14729163 87.0 8e-18
SPBC24C6.03 86.3 1e-17
7292265 82.8 1e-16
YER087w 75.9 2e-14
CE16514 72.4 2e-13
7294319 40.8 6e-04
Hs6996010 40.4 8e-04
At1g29870 40.0 0.001
YBR121c 39.7 0.001
At1g29880 39.7 0.001
YPR081c 38.9 0.003
CE29472 38.1 0.004
SPAC3F10.03 37.7 0.005
SPBC19C7.06 36.2 0.015
ECU10g1790 35.0 0.033
YHR020w 31.2 0.43
At5g52520 31.2 0.52
Hs17457197 30.8 0.70
ECU02g1360 30.4 0.86
Hs4758294_2 30.4 0.90
At3g62120 30.0 1.2
At5g26830 29.3 1.9
7301076_2 28.5 3.2
YER093c 27.7 5.1
At5g58260 27.3 7.0
Hs20270046 27.3 7.7
> Hs14729163
Length=402
Score = 87.0 bits (214), Expect = 8e-18, Method: Composition-based stats.
Identities = 42/96 (43%), Positives = 62/96 (64%), Gaps = 1/96 (1%)
Query 7 LSLPNIMPIHWLQNSDRIRSFGPSLYRLQDRRGRQFFLPPTSEEAAAWLVASNIR-SHRD 65
+++P++ P Q ++R G L RL+DR G+++ L PT EEA L+AS + S++
Sbjct 44 VNMPSLSPAELWQATNRWDLMGKELLRLRDRHGKEYCLGPTHEEAITALIASQKKLSYKQ 103
Query 66 LPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
LP + YQ+ KFRDE RPR G LR REF M ++Y+F
Sbjct 104 LPFLLYQVTRKFRDEPRPRFGLLRGREFYMKDMYTF 139
> SPBC24C6.03
Length=425
Score = 86.3 bits (212), Expect = 1e-17, Method: Composition-based stats.
Identities = 44/98 (44%), Positives = 58/98 (59%), Gaps = 0/98 (0%)
Query 4 ASPLSLPNIMPIHWLQNSDRIRSFGPSLYRLQDRRGRQFFLPPTSEEAAAWLVASNIRSH 63
AS +SL ++ + S R + G L+RL DR R+ L PT EE +A+ I S
Sbjct 71 ASAISLAHLSSKEIWEKSGRWQKTGSELFRLHDRNDREMCLAPTHEEDVTRTMATIIDSQ 130
Query 64 RDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
+ LP YQIG KFRDE RPR G LR REF+M ++Y+F
Sbjct 131 KQLPIRVYQIGRKFRDELRPRGGLLRGREFMMKDLYTF 168
> 7292265
Length=458
Score = 82.8 bits (203), Expect = 1e-16, Method: Composition-based stats.
Identities = 40/96 (41%), Positives = 60/96 (62%), Gaps = 1/96 (1%)
Query 7 LSLPNIMPIHWLQNSDRIRSFGPSLYRLQDRRGRQFFLPPTSEEA-AAWLVASNIRSHRD 65
++LP + P + + R+ Y ++DR G+QF + PT EEA A L ++ S+R
Sbjct 76 ITLPILTPTGLWKKTGRLDGDISEFYMVRDRSGKQFLMSPTHEEAVTAMLATTSPISYRQ 135
Query 66 LPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
LP +QIGPKFRDE + R G +RA+EF+M ++YSF
Sbjct 136 LPLRLFQIGPKFRDELKTRFGLMRAKEFLMKDMYSF 171
> YER087w
Length=576
Score = 75.9 bits (185), Expect = 2e-14, Method: Composition-based stats.
Identities = 40/98 (40%), Positives = 58/98 (59%), Gaps = 2/98 (2%)
Query 4 ASPLSLPNIMPIHWLQNSDRIRSFGPSLYRLQDRRGRQFFLPPTSEEAAAWLVASNIRSH 63
A +SL I Q +DR + L++L+D +G+Q+ L T EE L+ + I S+
Sbjct 74 AIEVSLSAISSKALWQATDRWNN--SELFKLKDSKGKQYCLTATCEEDITDLMKNYIASY 131
Query 64 RDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
+D+P YQ+ K+RDE RPR G LR REF+M + YSF
Sbjct 132 KDMPITIYQMTRKYRDEIRPRGGILRGREFLMKDAYSF 169
> CE16514
Length=454
Score = 72.4 bits (176), Expect = 2e-13, Method: Composition-based stats.
Identities = 36/99 (36%), Positives = 54/99 (54%), Gaps = 1/99 (1%)
Query 4 ASPLSLPNIMPIHWLQNSDRIRSFGPSLYRLQDRRGRQFFLPPTSEEAAAWLVAS-NIRS 62
A +++P + + + R + GP + R DR+ L PT+EE L+A+ +
Sbjct 66 ALKIAMPIVGTQQLWEKTGRWEAMGPEMIRFHDRQQNHMCLQPTAEEMCTELIATLSPLK 125
Query 63 HRDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
P I YQIG KFRDE PR G +R R+F+M ++YSF
Sbjct 126 KSQFPLIVYQIGEKFRDEMNPRFGLMRGRQFLMKDMYSF 164
> 7294319
Length=765
Score = 40.8 bits (94), Expect = 6e-04, Method: Composition-based stats.
Identities = 18/36 (50%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 66 LPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
LP QIG FR+E PR+G +R REF M E+ F
Sbjct 351 LPFAVAQIGNSFRNEISPRSGLIRVREFTMAEIEHF 386
> Hs6996010
Length=685
Score = 40.4 bits (93), Expect = 8e-04, Method: Composition-based stats.
Identities = 18/36 (50%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 66 LPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
LP QIG FR+E PR+G +R REF M E+ F
Sbjct 265 LPFAAAQIGNSFRNEISPRSGLIRVREFTMAEIEHF 300
> At1g29870
Length=492
Score = 40.0 bits (92), Expect = 0.001, Method: Composition-based stats.
Identities = 18/38 (47%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 64 RDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
R LP Q+G FR+E PR G LR REF + E+ F
Sbjct 257 RKLPFAVAQVGRVFRNEISPRQGLLRTREFTLAEIEHF 294
> YBR121c
Length=667
Score = 39.7 bits (91), Expect = 0.001, Method: Composition-based stats.
Identities = 17/29 (58%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 73 IGPKFRDEARPRAGCLRAREFVMLEVYSF 101
IG FR+E PRAG LR REF+M E+ F
Sbjct 255 IGKSFRNEISPRAGLLRVREFLMAEIEHF 283
> At1g29880
Length=690
Score = 39.7 bits (91), Expect = 0.001, Method: Composition-based stats.
Identities = 18/38 (47%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 64 RDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
+ LP QIG FR+E PR G LR REF + E+ F
Sbjct 275 KKLPFAAAQIGQAFRNEISPRQGLLRVREFTLAEIEHF 312
> YPR081c
Length=618
Score = 38.9 bits (89), Expect = 0.003, Method: Composition-based stats.
Identities = 17/36 (47%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 66 LPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
+P IG FR+E PR+G LR REF+M E+ F
Sbjct 207 IPFASASIGKSFRNEISPRSGLLRVREFLMAEIEHF 242
> CE29472
Length=742
Score = 38.1 bits (87), Expect = 0.004, Method: Composition-based stats.
Identities = 18/36 (50%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 66 LPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
LP QIG FR+E PR G +R REF M E+ F
Sbjct 329 LPFAAAQIGLGFRNEISPRQGLIRVREFTMCEIEHF 364
> SPAC3F10.03
Length=652
Score = 37.7 bits (86), Expect = 0.005, Method: Composition-based stats.
Identities = 16/29 (55%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 73 IGPKFRDEARPRAGCLRAREFVMLEVYSF 101
+G FR+E PR+G LR REF+M EV F
Sbjct 248 VGKAFRNEISPRSGLLRVREFLMAEVEHF 276
> SPBC19C7.06
Length=716
Score = 36.2 bits (82), Expect = 0.015, Method: Composition-based stats.
Identities = 27/88 (30%), Positives = 36/88 (40%), Gaps = 4/88 (4%)
Query 19 QNSDRIRSFGPSLYRLQDRRGRQFFLP----PTSEEAAAWLVASNIRSHRDLPQIFYQIG 74
+ D + F P + + + P PTSE A IRSHRDLP Q
Sbjct 280 KEKDHVEGFAPEVAWVTRAGTSELDEPIAIRPTSETVMYPYYAKWIRSHRDLPLKLNQWN 339
Query 75 PKFRDEARPRAGCLRAREFVMLEVYSFH 102
R E + LR REF+ E ++ H
Sbjct 340 SVVRWEFKNPQPFLRTREFLWQEGHTAH 367
> ECU10g1790
Length=579
Score = 35.0 bits (79), Expect = 0.033, Method: Composition-based stats.
Identities = 21/64 (32%), Positives = 29/64 (45%), Gaps = 1/64 (1%)
Query 39 GRQFFLPPTSEEAAAWLVASNIRSHRD-LPQIFYQIGPKFRDEARPRAGCLRAREFVMLE 97
G+ FL P + + + D LP IG +R+E PR+G LR REF E
Sbjct 167 GQTMFLRPETAQGQFLNFKKLCEYNNDKLPFASASIGKAYRNEISPRSGLLRVREFDQAE 226
Query 98 VYSF 101
+ F
Sbjct 227 IEHF 230
> YHR020w
Length=688
Score = 31.2 bits (69), Expect = 0.43, Method: Composition-based stats.
Identities = 24/88 (27%), Positives = 36/88 (40%), Gaps = 4/88 (4%)
Query 19 QNSDRIRSFGPSLYRLQDRRGRQFFLP----PTSEEAAAWLVASNIRSHRDLPQIFYQIG 74
+ D + F P + + + P PTSE A ++S+RDLP Q
Sbjct 260 KEKDHVEGFAPEVAWVTRAGSSELEEPIAIRPTSETVMYPYYAKWVQSYRDLPLKLNQWN 319
Query 75 PKFRDEARPRAGCLRAREFVMLEVYSFH 102
R E + LR REF+ E ++ H
Sbjct 320 SVVRWEFKHPQPFLRTREFLWQEGHTAH 347
> At5g52520
Length=543
Score = 31.2 bits (69), Expect = 0.52, Method: Composition-based stats.
Identities = 26/103 (25%), Positives = 45/103 (43%), Gaps = 6/103 (5%)
Query 5 SPLSLPNIMPIHWLQN-SDRIRSFGPSLYRLQDRRGRQF----FLPPTSEEAAAWLVASN 59
S + P +P +++ + + F P L + G++ + PTSE +
Sbjct 123 SNMYFPQFIPYSFIEKEASHVEGFSPELALVTVGGGKELEEKLVVRPTSETIVNHMFTQW 182
Query 60 IRSHRDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSFH 102
I S+RDLP + Q R E R + +R EF+ E ++ H
Sbjct 183 IHSYRDLPLMINQWANVTRWEMRTKP-FIRTLEFLWQEGHTAH 224
> Hs17457197
Length=167
Score = 30.8 bits (68), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 34/69 (49%), Gaps = 1/69 (1%)
Query 1 REMASPLSLPNIMPIHWLQNSDRIRSFGPSLYRLQDRRGRQFFLPPTSEEAAAWLVASNI 60
R SP L +PI +L+N I G + + D +GR F + P + E A + V I
Sbjct 30 RPSTSPHKLRECLPIIFLRNRLNIDKTGENFRLIYDTKGR-FAVHPITPEEAKYKVNDTI 88
Query 61 RSHRDLPQI 69
++ ++ +I
Sbjct 89 QTDLEIGKI 97
> ECU02g1360
Length=520
Score = 30.4 bits (67), Expect = 0.86, Method: Composition-based stats.
Identities = 27/95 (28%), Positives = 40/95 (42%), Gaps = 7/95 (7%)
Query 9 LPNIMPIHWLQ-NSDRIRSFGPSLYRLQDRRGRQFF-----LPPTSEEAAAWLVASNIRS 62
P ++P L+ D + +F P + + + G Q + PTSE + IRS
Sbjct 89 FPMLVPKSMLEMEKDHVENFSPEVAWIT-KCGNQVLEDPVAVRPTSETIIYPSFSKWIRS 147
Query 63 HRDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLE 97
HRDLP Q R E +R +EF+ E
Sbjct 148 HRDLPLKLNQWCSVLRWELHGTLPFIRGKEFLWQE 182
> Hs4758294_2
Length=499
Score = 30.4 bits (67), Expect = 0.90, Method: Composition-based stats.
Identities = 21/55 (38%), Positives = 26/55 (47%), Gaps = 0/55 (0%)
Query 46 PTSEEAAAWLVASNIRSHRDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYS 100
PTSE A ++SHRDLP Q R E + LR REF+ E +S
Sbjct 107 PTSETVMYPAYAKWVQSHRDLPIKLNQWCNVVRWEFKHPQPFLRTREFLWQEGHS 161
> At3g62120
Length=530
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 24/83 (28%), Positives = 32/83 (38%), Gaps = 4/83 (4%)
Query 19 QNSDRIRSFGPSLYRLQDRRGRQFFLP----PTSEEAAAWLVASNIRSHRDLPQIFYQIG 74
+ D I F P + + +P PTSE + IR HRDLP Q
Sbjct 122 KEKDHIEGFAPEVAWVTKSGKSDLEVPIAIRPTSETVMYPYYSKWIRGHRDLPLKLNQWC 181
Query 75 PKFRDEARPRAGCLRAREFVMLE 97
R E +R+REF+ E
Sbjct 182 NVVRWEFSNPTPFIRSREFLWQE 204
> At5g26830
Length=676
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 21/93 (22%), Positives = 40/93 (43%), Gaps = 2/93 (2%)
Query 10 PNIMPIHWLQNSDRIRSFGPSLYRLQDRRGRQFFLPPTSEEAAAWLVASNIRSHRDLPQI 69
PN+ + Q S ++ +++ + ++F L P + + +RS+R+LP
Sbjct 336 PNMYNMELWQTSGHADNYKDNMFTFNIEK-QEFGLKPMNCPGHCLIFQHRVRSYRELPMR 394
Query 70 FYQIGPKFRDEAR-PRAGCLRAREFVMLEVYSF 101
G R+EA +G R R F + + F
Sbjct 395 LADFGVLHRNEASGALSGLTRVRRFQQDDAHIF 427
> 7301076_2
Length=586
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 19/52 (36%), Positives = 24/52 (46%), Gaps = 0/52 (0%)
Query 46 PTSEEAAAWLVASNIRSHRDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLE 97
PTSE A ++S+RDLP Q R E + LR REF+ E
Sbjct 193 PTSETVMYPAYAKWVQSYRDLPIRLNQWNNVVRWEFKQPTPFLRTREFLWQE 244
> YER093c
Length=1430
Score = 27.7 bits (60), Expect = 5.1, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 30/64 (46%), Gaps = 6/64 (9%)
Query 2 EMASPLSLPNIMPIHWLQNSDRIRSFGPSLYRLQDRRGRQFFLPPTSEEAAAWLVASNIR 61
E S LS+P+ L S+++ G + L D T+ EAA WLV+ ++
Sbjct 259 ENHSTLSIPD------LDGSNKVNLTGDTEKDLGDLENENQIFTSTTTEAATWLVSDYMQ 312
Query 62 SHRD 65
S ++
Sbjct 313 SFQE 316
> At5g58260
Length=209
Score = 27.3 bits (59), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 25/51 (49%), Gaps = 2/51 (3%)
Query 44 LPPTSEEAAAWLVASNIRSHRDLPQIFYQIGPKFRDEARPRAGCLRAREFV 94
LPP+++ W++ + + S +L F + P R R A C R+FV
Sbjct 147 LPPSAKGLVVWVLEAKVLSKSELQ--FLALLPSLRPNVRVIAECGNWRKFV 195
> Hs20270046
Length=4169
Score = 27.3 bits (59), Expect = 7.7, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 16/29 (55%), Gaps = 0/29 (0%)
Query 46 PTSEEAAAWLVASNIRSHRDLPQIFYQIG 74
P ++ A V S+ SHR P IFYQ G
Sbjct 561 PADQKTATPTVLSSSHSHRGKPSIFYQQG 589
Lambda K H
0.327 0.139 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1181107380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40