bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2155_orf1
Length=131
Score E
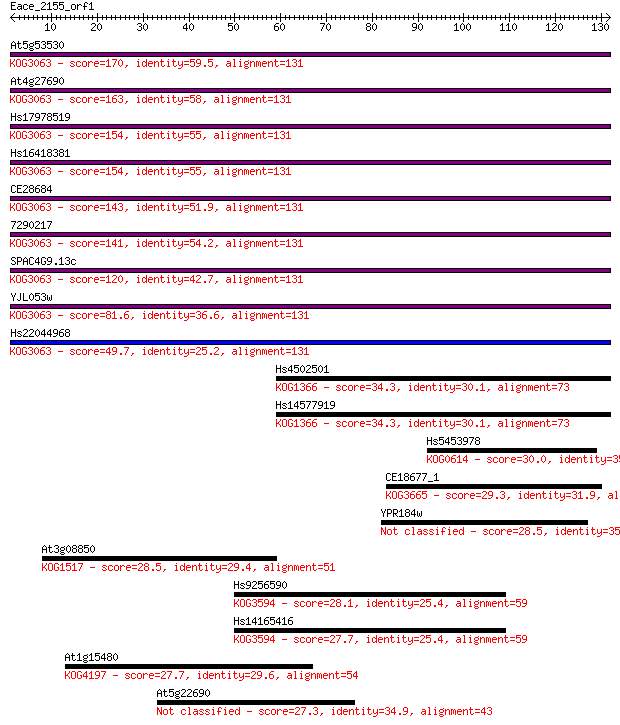
Sequences producing significant alignments: (Bits) Value
At5g53530 170 5e-43
At4g27690 163 9e-41
Hs17978519 154 3e-38
Hs16418381 154 5e-38
CE28684 143 7e-35
7290217 141 4e-34
SPAC4G9.13c 120 5e-28
YJL053w 81.6 4e-16
Hs22044968 49.7 2e-06
Hs4502501 34.3 0.069
Hs14577919 34.3 0.069
Hs5453978 30.0 1.3
CE18677_1 29.3 2.2
YPR184w 28.5 3.6
At3g08850 28.5 3.8
Hs9256590 28.1 5.0
Hs14165416 27.7 5.9
At1g15480 27.7 6.6
At5g22690 27.3 7.8
> At5g53530
Length=302
Score = 170 bits (431), Expect = 5e-43, Method: Compositional matrix adjust.
Identities = 78/131 (59%), Positives = 107/131 (81%), Gaps = 0/131 (0%)
Query 1 RSSTYDFFSITKDLDPPGVLVESKKYKFKFNAVDKTHETYSGINVRLRYFVRLTIHRHYA 60
R + YDF S+ +++D PG + E K Y F+F++V+ +ETY+G+NVRLRY +++T+ R YA
Sbjct 81 RGNFYDFTSLVREIDVPGEIYERKTYPFEFSSVEMPYETYNGVNVRLRYVLKVTVTRGYA 140
Query 61 SSIVKEHDFIVQNVGPPPEIKNSIKMEVGIEDCLHIEFEFDKSRYHLKDVVIGKVYFVLV 120
SIV+ DF+V+N P P I NSIKMEVGIEDCLHIEFE++KS+YHLKDV++GK+YF+LV
Sbjct 141 GSIVEYQDFVVRNYVPLPPINNSIKMEVGIEDCLHIEFEYNKSKYHLKDVILGKIYFLLV 200
Query 121 RIKIKDMQLDI 131
RIKIK+M L+I
Sbjct 201 RIKIKNMDLEI 211
> At4g27690
Length=302
Score = 163 bits (412), Expect = 9e-41, Method: Compositional matrix adjust.
Identities = 76/131 (58%), Positives = 105/131 (80%), Gaps = 1/131 (0%)
Query 1 RSSTYDFFSITKDLDPPGVLVESKKYKFKFNAVDKTHETYSGINVRLRYFVRLTIHRHYA 60
R + YDF S+ ++LD PG + E K Y F+F V+ +ETY+G+NVRLRY +++T+ R YA
Sbjct 81 RGNFYDFTSLVRELDVPGEIYERKTYPFEFPTVEMPYETYNGVNVRLRYVLKVTVTRGYA 140
Query 61 SSIVKEHDFIVQNVGPPPEIKNSIKMEVGIEDCLHIEFEFDKSRYHLKDVVIGKVYFVLV 120
SI+ E+ +V+N P P+I NSIKMEVGIEDCLHIEFE++KS+YHLKDV++GK+YF+LV
Sbjct 141 GSIL-EYQELVRNYAPLPDINNSIKMEVGIEDCLHIEFEYNKSKYHLKDVILGKIYFLLV 199
Query 121 RIKIKDMQLDI 131
RIK+K+M L+I
Sbjct 200 RIKMKNMDLEI 210
> Hs17978519
Length=327
Score = 154 bits (390), Expect = 3e-38, Method: Compositional matrix adjust.
Identities = 72/131 (54%), Positives = 104/131 (79%), Gaps = 1/131 (0%)
Query 1 RSSTYDFFSITKDLDPPGVLVESKKYKFKFNAVDKTHETYSGINVRLRYFVRLTIHRHYA 60
+S+T++F ++ K+L PG L +S+ Y F+F V+K +E+Y G NVRLRYF+++TI R
Sbjct 82 KSNTHEFVNLVKELALPGELTQSRSYDFEFMQVEKPYESYIGANVRLRYFLKVTIVRRL- 140
Query 61 SSIVKEHDFIVQNVGPPPEIKNSIKMEVGIEDCLHIEFEFDKSRYHLKDVVIGKVYFVLV 120
+ +VKE+D IV + P++ NSIKMEVGIEDCLHIEFE++KS+YHLKDV++GK+YF+LV
Sbjct 141 TDLVKEYDLIVHQLATYPDVNNSIKMEVGIEDCLHIEFEYNKSKYHLKDVIVGKIYFLLV 200
Query 121 RIKIKDMQLDI 131
RIKI+ M+L +
Sbjct 201 RIKIQHMELQL 211
> Hs16418381
Length=336
Score = 154 bits (388), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 72/131 (54%), Positives = 102/131 (77%), Gaps = 1/131 (0%)
Query 1 RSSTYDFFSITKDLDPPGVLVESKKYKFKFNAVDKTHETYSGINVRLRYFVRLTIHRHYA 60
R + ++F S+ KDL PG + +S+ + F+F V+K +E+Y+G NV+LRYF+R TI R
Sbjct 80 RGNHHEFVSLVKDLARPGEITQSQAFDFEFTHVEKPYESYTGQNVKLRYFLRATISRRL- 138
Query 61 SSIVKEHDFIVQNVGPPPEIKNSIKMEVGIEDCLHIEFEFDKSRYHLKDVVIGKVYFVLV 120
+ +VKE D +V + PE+ +SIKMEVGIEDCLHIEFE++KS+YHLKDV++GK+YF+LV
Sbjct 139 NDVVKEMDIVVHTLSTYPELNSSIKMEVGIEDCLHIEFEYNKSKYHLKDVIVGKIYFLLV 198
Query 121 RIKIKDMQLDI 131
RIKIK M++DI
Sbjct 199 RIKIKHMEIDI 209
> CE28684
Length=408
Score = 143 bits (361), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 68/131 (51%), Positives = 98/131 (74%), Gaps = 1/131 (0%)
Query 1 RSSTYDFFSITKDLDPPGVLVESKKYKFKFNAVDKTHETYSGINVRLRYFVRLTIHRHYA 60
R + DF S+T++L PG L ++ ++ F+FN V+K ETY G NV+LRYF+R+T+ R
Sbjct 133 RGNQQDFISLTRELARPGDLTQNAQFPFEFNNVEKPFETYMGTNVKLRYFLRVTVIRRL- 191
Query 61 SSIVKEHDFIVQNVGPPPEIKNSIKMEVGIEDCLHIEFEFDKSRYHLKDVVIGKVYFVLV 120
+ + KE D +V + P+ SIKMEVGIEDCLHIEFE++K++YHL+DV++GK+YF+LV
Sbjct 192 TDLTKELDLVVHALSSYPDNDKSIKMEVGIEDCLHIEFEYNKNKYHLQDVIVGKIYFLLV 251
Query 121 RIKIKDMQLDI 131
RIKIK M++ I
Sbjct 252 RIKIKYMEIAI 262
> 7290217
Length=478
Score = 141 bits (355), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 71/131 (54%), Positives = 92/131 (70%), Gaps = 1/131 (0%)
Query 1 RSSTYDFFSITKDLDPPGVLVESKKYKFKFNAVDKTHETYSGINVRLRYFVRLTIHRHYA 60
R + ++F + K L PG L+++ Y F F V+K E Y+G NVRLRYF+R TI R
Sbjct 79 RGNHHEFKCLAKALARPGDLIQNNSYPFDFPKVEKQFEVYAGSNVRLRYFLRATIVRRI- 137
Query 61 SSIVKEHDFIVQNVGPPPEIKNSIKMEVGIEDCLHIEFEFDKSRYHLKDVVIGKVYFVLV 120
S I KE D V + PE+ N IKMEVGIEDCLHIEFE++KS+YHL+D +IGK+YF+LV
Sbjct 138 SDITKEVDIAVHTLCSYPEMNNPIKMEVGIEDCLHIEFEYNKSKYHLRDTIIGKIYFLLV 197
Query 121 RIKIKDMQLDI 131
RIKIK M++ I
Sbjct 198 RIKIKHMEIAI 208
> SPAC4G9.13c
Length=298
Score = 120 bits (302), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 56/131 (42%), Positives = 91/131 (69%), Gaps = 1/131 (0%)
Query 1 RSSTYDFFSITKDLDPPGVLVESKKYKFKFNAVDKTHETYSGINVRLRYFVRLTIHRHYA 60
+ + ++F ++L PG + ++ ++F+F VDK +E+Y G NV+LRY R+T+ R
Sbjct 78 KGNIHEFTRSVQELASPGEMRHAQMFEFEFKHVDKPYESYIGKNVKLRYICRVTVSRK-M 136
Query 61 SSIVKEHDFIVQNVGPPPEIKNSIKMEVGIEDCLHIEFEFDKSRYHLKDVVIGKVYFVLV 120
+++E D V PE + I+M+VGI++CLHIEFE+ K++YHLKDV+IGK+YF+LV
Sbjct 137 KDVIREKDLWVYRFENEPETNSLIRMDVGIDECLHIEFEYSKNKYHLKDVIIGKIYFILV 196
Query 121 RIKIKDMQLDI 131
RIK++ M++ I
Sbjct 197 RIKVQRMEVSI 207
> YJL053w
Length=379
Score = 81.6 bits (200), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 48/184 (26%), Positives = 85/184 (46%), Gaps = 53/184 (28%)
Query 1 RSSTYDFFSITKDLDPPGVLVESKKYKFKFNAVDKTHETYSGINVRLRYFVRLTIHR--- 57
++S F + DL P G L S+ + F F + K +E+Y G NV + Y+V++T+ R
Sbjct 111 KNSVDQFLCQSYDLCPAGELQHSQSFPFLFRDLSKRYESYKGKNVDVAYYVKVTVMRKST 170
Query 58 --------------------------------------HYASSIVKEHDFIVQNVGPPPE 79
+YA+ ++ G +
Sbjct 171 DISKIKRFWVYLYNSVTTAPNTLSANETKATTNDIAGGNYAADNASDNTQTKSTQGEAAD 230
Query 80 IKN------------SIKMEVGIEDCLHIEFEFDKSRYHLKDVVIGKVYFVLVRIKIKDM 127
+ +++++GIE+CLHIEFE+ KS+Y LK+V++G++YF+L R++IK M
Sbjct 231 VNQVLPISHSNNEPKPVRLDIGIENCLHIEFEYAKSQYSLKEVIVGRIYFLLTRLRIKHM 290
Query 128 QLDI 131
+L +
Sbjct 291 ELSL 294
> Hs22044968
Length=120
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/131 (25%), Positives = 55/131 (41%), Gaps = 53/131 (40%)
Query 1 RSSTYDFFSITKDLDPPGVLVESKKYKFKFNAVDKTHETYSGINVRLRYFVRLTIHRHYA 60
+S+ ++F ++ K+L PG L + + Y F+F V K + + G +V L
Sbjct 20 KSNAHEFINLMKELALPGELTQRRSYDFEFMQVKKPYASDIGASVHL------------- 66
Query 61 SSIVKEHDFIVQNVGPPPEIKNSIKMEVGIEDCLHIEFEFDKSRYHLKDVVIGKVYFVLV 120
KS+YHLK V++GK+YF++V
Sbjct 67 ----------------------------------------SKSKYHLKHVIVGKIYFLVV 86
Query 121 RIKIKDMQLDI 131
RIKI+ M+L +
Sbjct 87 RIKIQHMELQL 97
> Hs4502501
Length=1744
Score = 34.3 bits (77), Expect = 0.069, Method: Composition-based stats.
Identities = 22/76 (28%), Positives = 37/76 (48%), Gaps = 4/76 (5%)
Query 59 YASSIVKEHDFIVQNVGPPPEIKNSIKMEVGIEDCLHIEFEFDK---SRYHLKDVVIGKV 115
Y S + + DF++ ++ P K S + G+E +FE K + +K + GK
Sbjct 192 YMPSSIFQDDFVIPDISEPGTWKISARFSDGLESNSSTQFEVKKYVLPNFEVK-ITPGKP 250
Query 116 YFVLVRIKIKDMQLDI 131
Y + V + +MQLDI
Sbjct 251 YILTVPGHLDEMQLDI 266
> Hs14577919
Length=1741
Score = 34.3 bits (77), Expect = 0.069, Method: Composition-based stats.
Identities = 22/76 (28%), Positives = 37/76 (48%), Gaps = 4/76 (5%)
Query 59 YASSIVKEHDFIVQNVGPPPEIKNSIKMEVGIEDCLHIEFEFDK---SRYHLKDVVIGKV 115
Y S + + DF++ ++ P K S + G+E +FE K + +K + GK
Sbjct 192 YMPSSIFQDDFVIPDISEPGTWKISARFSDGLESNSSTQFEVKKYVLPNFEVK-ITPGKP 250
Query 116 YFVLVRIKIKDMQLDI 131
Y + V + +MQLDI
Sbjct 251 YILTVPGHLDEMQLDI 266
> Hs5453978
Length=762
Score = 30.0 bits (66), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 13/37 (35%), Positives = 23/37 (62%), Gaps = 1/37 (2%)
Query 92 DCLHIEFEFDKSRYHLKDVVIGKVYFVLVRIKIKDMQ 128
DCL +E+ +DK Y +++ G +F+L + K+K Q
Sbjct 300 DCLEVEY-YDKGDYIIREGEEGSTFFILAKGKVKVTQ 335
> CE18677_1
Length=631
Score = 29.3 bits (64), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 83 SIKMEVGIEDCLHIEFEFDKSRYHLKDVVIGKVYFVLVRIKIKDMQL 129
++K +V + CLH F F KS YH+ + I VY + ++Q+
Sbjct 314 TLKHDVFMSQCLHALFNFIKSSYHIDNEWIQYVYRAINTCSSDNVQI 360
> YPR184w
Length=1536
Score = 28.5 bits (62), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 27/51 (52%), Gaps = 6/51 (11%)
Query 82 NSIKMEVG-----IEDC-LHIEFEFDKSRYHLKDVVIGKVYFVLVRIKIKD 126
NSI+ +G IED +H+ + +H DV GK ++++ R+K D
Sbjct 741 NSIRTSIGEKAYDIEDSEMHVHHQGQYITFHRMDVKSGKGWYLIARMKFSD 791
> At3g08850
Length=1384
Score = 28.5 bits (62), Expect = 3.8, Method: Composition-based stats.
Identities = 15/51 (29%), Positives = 22/51 (43%), Gaps = 0/51 (0%)
Query 8 FSITKDLDPPGVLVESKKYKFKFNAVDKTHETYSGINVRLRYFVRLTIHRH 58
S LDP V+ S+ +F + T +TY I+ L +HRH
Sbjct 1262 LSFQPGLDPAKVVSASQAGDIQFLDLRTTRDTYLTIDAHRGSLTALAVHRH 1312
> Hs9256590
Length=936
Score = 28.1 bits (61), Expect = 5.0, Method: Composition-based stats.
Identities = 15/60 (25%), Positives = 30/60 (50%), Gaps = 1/60 (1%)
Query 50 FVRLTIHRHYASSIVKEHDFIV-QNVGPPPEIKNSIKMEVGIEDCLHIEFEFDKSRYHLK 108
F+ L + + +EH +V G PE+ ++++ + + D EFDKS Y+++
Sbjct 190 FLELVLRKSLDREETQEHRLLVIATDGGKPELTGTVQLLINVLDANDNAPEFDKSIYNVR 249
> Hs14165416
Length=816
Score = 27.7 bits (60), Expect = 5.9, Method: Composition-based stats.
Identities = 15/60 (25%), Positives = 30/60 (50%), Gaps = 1/60 (1%)
Query 50 FVRLTIHRHYASSIVKEHDFIV-QNVGPPPEIKNSIKMEVGIEDCLHIEFEFDKSRYHLK 108
F+ L + + +EH +V G PE+ ++++ + + D EFDKS Y+++
Sbjct 190 FLELVLRKSLDREETQEHRLLVIATDGGKPELTGTVQLLINVLDANDNAPEFDKSIYNVR 249
> At1g15480
Length=623
Score = 27.7 bits (60), Expect = 6.6, Method: Composition-based stats.
Identities = 16/54 (29%), Positives = 27/54 (50%), Gaps = 0/54 (0%)
Query 13 DLDPPGVLVESKKYKFKFNAVDKTHETYSGINVRLRYFVRLTIHRHYASSIVKE 66
+L+ +L+++K +++ ET V L R I RHYAS+ +KE
Sbjct 324 NLNTYKILIDTKGSSNDITGMEQIVETMKSEGVELDLRARALIARHYASAGLKE 377
> At5g22690
Length=1008
Score = 27.3 bits (59), Expect = 7.8, Method: Composition-based stats.
Identities = 15/43 (34%), Positives = 23/43 (53%), Gaps = 1/43 (2%)
Query 33 VDKTHETYSGINVRLRYFVRLTIHRHYASSIVKEHDFIVQNVG 75
V K+ E YSG NV Y +L + + S I++ D + N+G
Sbjct 244 VSKSMEIYSGGNVD-NYNAKLHLQGKFLSEILRAKDIKISNLG 285
Lambda K H
0.324 0.142 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1283105810
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40