bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2115_orf1
Length=116
Score E
Sequences producing significant alignments: (Bits) Value
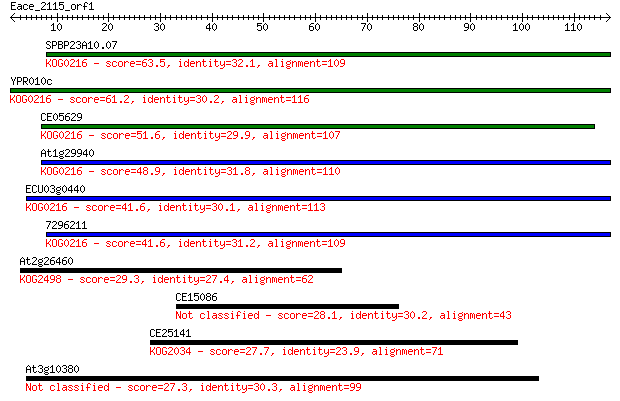
SPBP23A10.07 63.5 8e-11
YPR010c 61.2 5e-10
CE05629 51.6 3e-07
At1g29940 48.9 2e-06
ECU03g0440 41.6 3e-04
7296211 41.6 3e-04
At2g26460 29.3 2.1
CE15086 28.1 3.8
CE25141 27.7 5.6
At3g10380 27.3 6.8
> SPBP23A10.07
Length=1227
Score = 63.5 bits (153), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 35/110 (31%), Positives = 63/110 (57%), Gaps = 6/110 (5%)
Query 8 ESMDAFSYQEVVLPGQILASVLKDALFAALIKLRTQYMMELRQVKAAGHDPTLVISSIKF 67
++ D+ +QE++L G + +LK+ + L +R Q +++R+ P + S K+
Sbjct 420 DNPDSPQHQEILLGGFLYGQILKEKIEDWLNSIRAQINLDVRRSA-----PGVDFSDRKY 474
Query 68 FDAATERCASEIGHKINYFMATGN-IRTTQLDLQQLSGWTVTADRLNLNR 116
+ ++IG K+ YF++TGN + T LDLQQ +G+TV A++LN R
Sbjct 475 LTRVFSKINNDIGTKLQYFLSTGNLVSNTGLDLQQATGYTVVAEKLNFYR 524
> YPR010c
Length=1203
Score = 61.2 bits (147), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 35/117 (29%), Positives = 62/117 (52%), Gaps = 7/117 (5%)
Query 1 ICWKIPPESMDAFSYQEVVLPGQILASVLKDALFAALIKLRTQYMMELRQVKAAGHDPTL 60
+ + P++ DA +QEV+L G + +LK+ + L + Q M++ + A +
Sbjct 385 VAGECSPDNPDATQHQEVLLGGFLYGMILKEKIDEYLQNIIAQVRMDINRGMA------I 438
Query 61 VISSIKFFDAATERCASEIGHKINYFMATGN-IRTTQLDLQQLSGWTVTADRLNLNR 116
++ R IG K+ YF++TGN + + LDLQQ+SG+TV A+++N R
Sbjct 439 NFKDKRYMSRVLMRVNENIGSKMQYFLSTGNLVSQSGLDLQQVSGYTVVAEKINFYR 495
> CE05629
Length=1127
Score = 51.6 bits (122), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 32/110 (29%), Positives = 55/110 (50%), Gaps = 12/110 (10%)
Query 7 PESMDAFSYQEVVLPGQILASVLKDAL--FAALIKLRTQYMMELRQVKAAGHDPTLVISS 64
PE+ D +QE + G IL +L++ + +++ + +YM + +++S
Sbjct 335 PETPDNPQFQEASVSGHILLLILRERMENIIGMVRRKLEYMSSRKD---------FILTS 385
Query 65 IKFFDAATERCASEIGHKINYFMATGNIRT-TQLDLQQLSGWTVTADRLN 113
A EI + YF+ATGN+ T L LQQ SG++V A+R+N
Sbjct 386 AAILKALGNHTGGEITRGMAYFLATGNLVTRVGLALQQESGFSVIAERIN 435
> At1g29940
Length=1114
Score = 48.9 bits (115), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 35/111 (31%), Positives = 56/111 (50%), Gaps = 5/111 (4%)
Query 7 PESMDAFSYQEVVLPGQILASVLKDALFAALIKLRTQYMMELRQVKAAGHDPTLVISSIK 66
P++ D+ QE+++PG ++ LK+ L L K ++ EL + +L K
Sbjct 302 PDNPDSLQNQEILVPGHVITIYLKEKLEEWLRKCKSLLKDELDNTNSKFSFESLA-DVKK 360
Query 67 FFDAATERCASEIGHKINYFMATGNIRT-TQLDLQQLSGWTVTADRLNLNR 116
+ R IG I + TG ++T + LDLQQ +G+TV A+RLN R
Sbjct 361 LINKNPPR---SIGTSIETLLKTGALKTQSGLDLQQRAGYTVQAERLNFLR 408
> ECU03g0440
Length=1062
Score = 41.6 bits (96), Expect = 3e-04, Method: Composition-based stats.
Identities = 34/127 (26%), Positives = 64/127 (50%), Gaps = 17/127 (13%)
Query 4 KIPPESMDAFSYQEVVLPGQILASVLKDAL-----FAALIKLRTQYMMELRQVKAAGHDP 58
+I + D+ S E+ QI++S L+D + F +++ +T Y +E + K G
Sbjct 322 EITEDDPDSPSNHELGTETQIISSFLRDRIEEFMRFFSMMCRKTYYKLEGEKRKLEGDSV 381
Query 59 T--------LVISSIKFFDAATERCASEIGHKINYFMATGNIRTTQL-DLQQLSGWTVTA 109
T L IS I+ +R + IG ++ F+++G+I T D+ Q G+++TA
Sbjct 382 TESGFCEDELDISKIR---EIFKRTSINIGQRMELFLSSGSITLTSCSDILQTLGFSITA 438
Query 110 DRLNLNR 116
+R+N +
Sbjct 439 ERINFYK 445
> 7296211
Length=1129
Score = 41.6 bits (96), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 34/110 (30%), Positives = 54/110 (49%), Gaps = 10/110 (9%)
Query 8 ESMDAFSYQEVVLPGQILASVLKDALFAALIKLRTQYMMELRQVKAAGHDPTLVISSIKF 67
E++D+ QEV+LPG + L + + + + ++R +L A LV S++
Sbjct 364 ENVDSAMMQEVLLPGHLYQKYLSERVESWVSQVRRCLQKKLTSPDA------LVTSAVM- 416
Query 68 FDAATERCASEIGHKINYFMATGNIRT-TQLDLQQLSGWTVTADRLNLNR 116
R A +G I F+ATGNI + T L L Q SG + A+ +N R
Sbjct 417 --TQCMRQAGGVGRAIESFLATGNIASRTGLGLMQNSGLVIMAENINRMR 464
> At2g26460
Length=585
Score = 29.3 bits (64), Expect = 2.1, Method: Composition-based stats.
Identities = 17/62 (27%), Positives = 31/62 (50%), Gaps = 7/62 (11%)
Query 3 WKIPPESMDAFSYQEVVLPGQILASVLKDALFAALIKLRTQYMMELRQVKAAGHDPTLVI 62
+++ PE+ +V+L Q+++ KD ++ K Q + +LR+ A DPT V
Sbjct 406 YEVQPET-------DVLLDPQLMSQEEKDRGLGSVFKRDDQRLQQLRESDAREKDPTFVS 458
Query 63 SS 64
S
Sbjct 459 ES 460
> CE15086
Length=430
Score = 28.1 bits (61), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 13/43 (30%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 33 LFAALIKLRTQYMMELRQVKAAGHDPTLVISSIKFFDAATERC 75
L KL++Q E R+++ AG + V ++ + +AA +RC
Sbjct 279 LHPEQAKLKSQRWQETRRLRRAGLNQEKVEKALPYLEAAFKRC 321
> CE25141
Length=1010
Score = 27.7 bits (60), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 17/71 (23%), Positives = 31/71 (43%), Gaps = 11/71 (15%)
Query 28 VLKDALFAALIKLRTQYMMELRQVKAAGHDPTLVISSIKFFDAATERCASEIGHKINYFM 87
+ +DAL L+ ++ + + E+R++K + DP V E+ H YFM
Sbjct 488 IRRDALVMWLLNVQLEELAEMRRLKNSNPDPAFV-----------EKLRDTTDHVQRYFM 536
Query 88 ATGNIRTTQLD 98
I + Q +
Sbjct 537 RKNVIESIQTN 547
> At3g10380
Length=671
Score = 27.3 bits (59), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 30/102 (29%), Positives = 47/102 (46%), Gaps = 20/102 (19%)
Query 4 KIPPESMDAFSYQEVVLPGQ---ILASVLKDALFAALIKLRTQYMMELRQVKAAGHDPTL 60
+ P S++ + VLP Q + AS+ + L A L KL YM+ R HD
Sbjct 458 RAPNASLEGYG-SAAVLPEQGIYLAASIYRPVLQAVLEKL--SYMLIGR------HD--- 505
Query 61 VISSIKFFDAATERCASEIGHKINYFMATGNIRTTQLDLQQL 102
I + DAA+ S +GH +++ A G T+++L L
Sbjct 506 -IEKLMRLDAASACLPSTLGHAVSHSEAVG----TEVELSDL 542
Lambda K H
0.323 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174970866
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40