bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2056_orf1
Length=133
Score E
Sequences producing significant alignments: (Bits) Value
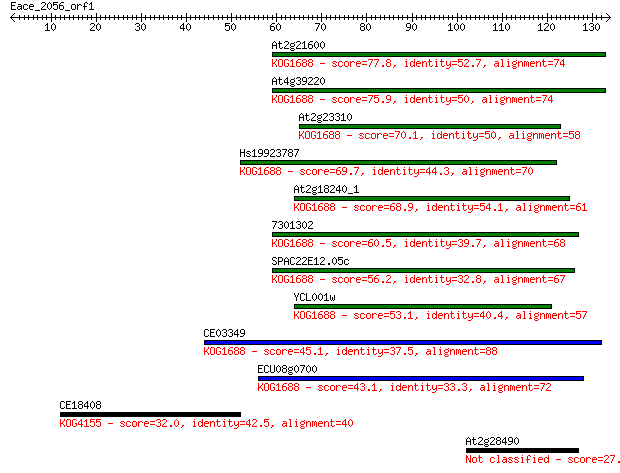
At2g21600 77.8 5e-15
At4g39220 75.9 2e-14
At2g23310 70.1 1e-12
Hs19923787 69.7 1e-12
At2g18240_1 68.9 2e-12
7301302 60.5 8e-10
SPAC22E12.05c 56.2 2e-08
YCL001w 53.1 1e-07
CE03349 45.1 3e-05
ECU08g0700 43.1 1e-04
CE18408 32.0 0.32
At2g28490 27.7 6.4
> At2g21600
Length=195
Score = 77.8 bits (190), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 39/77 (50%), Positives = 50/77 (64%), Gaps = 3/77 (3%)
Query 59 RVSSVYLNKSILYLKTRWLVLFACLAFYIVRVYFLAGFFVVSYGLGIYLLNLLIGFISPQ 118
RV YL+K+ + RW+ Y +RVY + GF+++SYGLGIYLLNLLIGF+SP
Sbjct 24 RVYQYYLDKTTPHSTNRWIGTLVFFLIYCLRVYSIHGFYIISYGLGIYLLNLLIGFLSPL 83
Query 119 VDPE---TDEYVLPVRG 132
VDPE +D LP RG
Sbjct 84 VDPELEVSDGATLPTRG 100
> At4g39220
Length=191
Score = 75.9 bits (185), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 37/78 (47%), Positives = 51/78 (65%), Gaps = 4/78 (5%)
Query 59 RVSSVYLNKSILYLKTRWLVLFACLAFYIVRVYFLAGFFVVSYGLGIYLLNLLIGFISPQ 118
R+ YL+K+ + RW+ Y +RVY++ GF++++YGLGIYLLNLLIGF+SP
Sbjct 24 RIYQHYLDKTTPHANYRWIGTLVVALIYCLRVYYIQGFYIIAYGLGIYLLNLLIGFLSPL 83
Query 119 VDPE----TDEYVLPVRG 132
VDPE +D LP RG
Sbjct 84 VDPEAGGVSDGPSLPTRG 101
> At2g23310
Length=211
Score = 70.1 bits (170), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 29/58 (50%), Positives = 44/58 (75%), Gaps = 0/58 (0%)
Query 65 LNKSILYLKTRWLVLFACLAFYIVRVYFLAGFFVVSYGLGIYLLNLLIGFISPQVDPE 122
L+K++ ++ RW+ + YIVRVYF+ GF++++Y +GIYLLNL+I F+SPQ DPE
Sbjct 52 LDKTVPHVLYRWIACLCVVLIYIVRVYFVEGFYIITYAIGIYLLNLIIAFLSPQEDPE 109
> Hs19923787
Length=206
Score = 69.7 bits (169), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 31/70 (44%), Positives = 46/70 (65%), Gaps = 0/70 (0%)
Query 52 RVWASVGRVSSVYLNKSILYLKTRWLVLFACLAFYIVRVYFLAGFFVVSYGLGIYLLNLL 111
R + +G++ +L+KS Y RW+V Y++RVY L G+++V+Y LGIY LNL
Sbjct 20 RFFTRLGQIYQSWLDKSTPYTAVRWVVTLGLSFVYMIRVYLLQGWYIVTYALGIYHLNLF 79
Query 112 IGFISPQVDP 121
I F+SP+VDP
Sbjct 80 IAFLSPKVDP 89
> At2g18240_1
Length=180
Score = 68.9 bits (167), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 33/61 (54%), Positives = 43/61 (70%), Gaps = 0/61 (0%)
Query 64 YLNKSILYLKTRWLVLFACLAFYIVRVYFLAGFFVVSYGLGIYLLNLLIGFISPQVDPET 123
YL++S + RWLV YI RVY + G+FV+SYGL Y+LNLLIGF+SP+VDPE
Sbjct 33 YLDRSAPNIVRRWLVTLVAAVIYIYRVYSVYGYFVISYGLATYILNLLIGFLSPKVDPEL 92
Query 124 D 124
+
Sbjct 93 E 93
> 7301302
Length=203
Score = 60.5 bits (145), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 27/71 (38%), Positives = 45/71 (63%), Gaps = 3/71 (4%)
Query 59 RVSSVY---LNKSILYLKTRWLVLFACLAFYIVRVYFLAGFFVVSYGLGIYLLNLLIGFI 115
R+S Y L++S + + RW+ L +++R++ G+++V Y LGIY LNL I F+
Sbjct 20 RLSQTYQSALDRSTPHTRMRWVFAGFLLLLFVLRIFIYQGWYIVCYALGIYHLNLFIAFL 79
Query 116 SPQVDPETDEY 126
+P++DPE D Y
Sbjct 80 TPKIDPEFDPY 90
> SPAC22E12.05c
Length=184
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 43/67 (64%), Gaps = 0/67 (0%)
Query 59 RVSSVYLNKSILYLKTRWLVLFACLAFYIVRVYFLAGFFVVSYGLGIYLLNLLIGFISPQ 118
R+ +++++I Y RWL + +A + +R+ + G+++V Y L IYLLNL + F++P+
Sbjct 20 RLYRHWVDRTIPYTTYRWLTVSGLIALFFIRILLVRGWYIVCYTLAIYLLNLFLAFLTPK 79
Query 119 VDPETDE 125
DP ++
Sbjct 80 FDPSVEQ 86
> YCL001w
Length=188
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 23/57 (40%), Positives = 37/57 (64%), Gaps = 0/57 (0%)
Query 64 YLNKSILYLKTRWLVLFACLAFYIVRVYFLAGFFVVSYGLGIYLLNLLIGFISPQVD 120
YL+K + K RW VL L ++VR+ G++V+ YGLG++LLN + F++P+ D
Sbjct 31 YLDKVTPHAKERWAVLGGLLCLFMVRITMAEGWYVICYGLGLFLLNQFLAFLTPKFD 87
> CE03349
Length=191
Score = 45.1 bits (105), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 33/95 (34%), Positives = 53/95 (55%), Gaps = 7/95 (7%)
Query 44 LADQP-FSSRVWASVGRVSSVYLNKSILYLKTRWLVLFACLAFYIVRVYFLAGFFVVSYG 102
L D+P +SR + S+ YL++ + RW++ L F+ R+ L GF++V+Y
Sbjct 5 LRDRPGVTSRFFHSLEVKYQYYLDRLTPHTAFRWVIALISLVFFASRIILLQGFYIVAYA 64
Query 103 LGIYLLNLLIGFISPQVDP------ETDEYVLPVR 131
+GIY LNL + F++P +DP E D VLP +
Sbjct 65 VGIYYLNLFLLFLTPSIDPALEFEDEDDGPVLPSK 99
> ECU08g0700
Length=166
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 24/80 (30%), Positives = 43/80 (53%), Gaps = 8/80 (10%)
Query 56 SVGRVSSVYLNKSILYLKTRWLVLFACLAFYIVRVYFLAGFFVVSYGLGIYLLNLLIGFI 115
+ + +YL++ RW + FY +R++ F++++Y LGIYLL+ LI F+
Sbjct 2 DLKTLQQIYLDRLAPRPDVRWGITGVLFLFYCIRIWSTGAFYLITYCLGIYLLHALILFL 61
Query 116 SPQ----VDP----ETDEYV 127
+P+ DP E D+Y+
Sbjct 62 TPKGEMIPDPFENIEDDDYI 81
> CE18408
Length=1101
Score = 32.0 bits (71), Expect = 0.32, Method: Composition-based stats.
Identities = 17/40 (42%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 12 SVGRGSERRRAAFAHSIESAVQTMMASMSAEELADQPFSS 51
+VG + RRR+A A S+ SA +AS SA E+ P S
Sbjct 1004 AVGVEATRRRSARAQSVSSASNEPVASTSAGEIKKDPILS 1043
> At2g28490
Length=511
Score = 27.7 bits (60), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 11/26 (42%), Positives = 18/26 (69%), Gaps = 1/26 (3%)
Query 102 GLGIYLLNLLIG-FISPQVDPETDEY 126
G+G+YL+NL G ++P ++P EY
Sbjct 362 GIGVYLVNLTAGAMMAPHMNPTATEY 387
Lambda K H
0.326 0.139 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1356426142
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40