bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
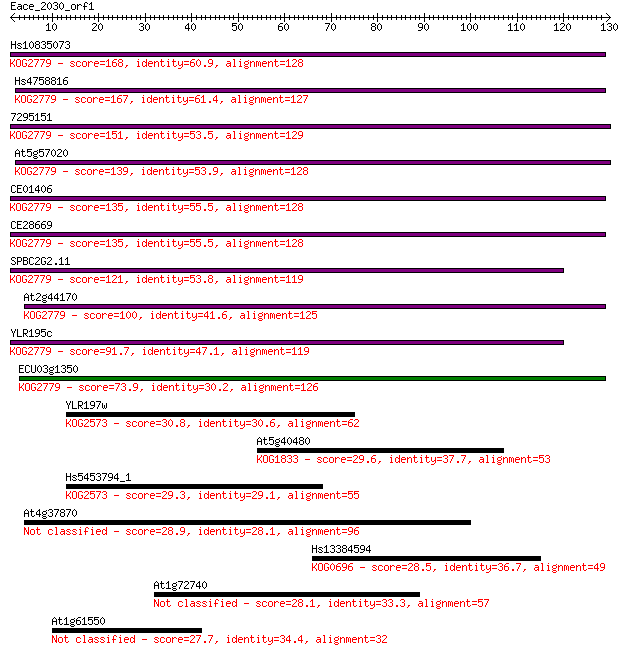
Query= Eace_2030_orf1
Length=129
Score E
Sequences producing significant alignments: (Bits) Value
Hs10835073 168 3e-42
Hs4758816 167 4e-42
7295151 151 3e-37
At5g57020 139 1e-33
CE01406 135 2e-32
CE28669 135 2e-32
SPBC2G2.11 121 3e-28
At2g44170 100 9e-22
YLR195c 91.7 3e-19
ECU03g1350 73.9 7e-14
YLR197w 30.8 0.66
At5g40480 29.6 1.5
Hs5453794_1 29.3 2.1
At4g37870 28.9 2.6
Hs13384594 28.5 3.2
At1g72740 28.1 4.2
At1g61550 27.7 5.2
> Hs10835073
Length=496
Score = 168 bits (425), Expect = 3e-42, Method: Compositional matrix adjust.
Identities = 78/129 (60%), Positives = 98/129 (75%), Gaps = 1/129 (0%)
Query 1 TQEEVRHWLMPKEGVVHVYVREQG-GKVTDLISFYELPSSVIGNQKHKEVKAAYSFYNVG 59
+QEEV HW P+E ++ +V E G+VTD +SFY LPS+++ + HK +KAAYSFYNV
Sbjct 367 SQEEVEHWFYPQENIIDTFVVENANGEVTDFLSFYTLPSTIMNHPTHKSLKAAYSFYNVH 426
Query 60 TSAPLKELIQDALCLAKQKDFDVFNALDVMENKTFVEDLKFGIGDGFLRYYLYNWRCPQV 119
T PL +L+ DAL LAK K FDVFNALD+MENKTF+E LKFGIGDG L+YYLYNW+CP +
Sbjct 427 TQTPLLDLMSDALVLAKMKGFDVFNALDLMENKTFLEKLKFGIGDGNLQYYLYNWKCPSM 486
Query 120 LHNDIGLVL 128
+GLVL
Sbjct 487 GAEKVGLVL 495
> Hs4758816
Length=498
Score = 167 bits (423), Expect = 4e-42, Method: Compositional matrix adjust.
Identities = 78/128 (60%), Positives = 97/128 (75%), Gaps = 1/128 (0%)
Query 2 QEEVRHWLMPKEGVVHVYVREQ-GGKVTDLISFYELPSSVIGNQKHKEVKAAYSFYNVGT 60
+EEV HW +P+E ++ +V E GK+TD +SFY LPS+V+ + HK +KAAYSFYN+ T
Sbjct 370 EEEVAHWFLPREHIIDTFVVESPNGKLTDFLSFYTLPSTVMHHPAHKSLKAAYSFYNIHT 429
Query 61 SAPLKELIQDALCLAKQKDFDVFNALDVMENKTFVEDLKFGIGDGFLRYYLYNWRCPQVL 120
PL +L+ DAL LAK K FDVFNALD+MENKTF+E LKFGIGDG L+YYLYNWRCP
Sbjct 430 ETPLLDLMSDALILAKSKGFDVFNALDLMENKTFLEKLKFGIGDGNLQYYLYNWRCPGTD 489
Query 121 HNDIGLVL 128
+GLVL
Sbjct 490 SEKVGLVL 497
> 7295151
Length=472
Score = 151 bits (382), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 69/130 (53%), Positives = 92/130 (70%), Gaps = 1/130 (0%)
Query 1 TQEEVRHWLMPKEGVVHVYV-REQGGKVTDLISFYELPSSVIGNQKHKEVKAAYSFYNVG 59
++EE RHW PKEG++ +V ++ G +TDL S+Y LPSSV+ + HK V+AAYSFYNV
Sbjct 343 SKEEFRHWFTPKEGIIDCFVVADEKGNITDLTSYYCLPSSVMHHPVHKTVRAAYSFYNVS 402
Query 60 TSAPLKELIQDALCLAKQKDFDVFNALDVMENKTFVEDLKFGIGDGFLRYYLYNWRCPQV 119
T P +L+ DAL A+ DV+NALD+MENK + LKFG GDG L+YYLYNWRCP +
Sbjct 403 TKTPWLDLMNDALISARNVQMDVYNALDLMENKKYFAPLKFGAGDGNLQYYLYNWRCPSM 462
Query 120 LHNDIGLVLL 129
+I L+L+
Sbjct 463 QPEEIALILM 472
> At5g57020
Length=370
Score = 139 bits (350), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 69/131 (52%), Positives = 93/131 (70%), Gaps = 3/131 (2%)
Query 2 QEEVRHWLMPKEGVVHVYVRE--QGGKVTDLISFYELPSSVIGNQKHKEVKAAYSFYNVG 59
+ +V HWL+P+E VV Y+ E + VTD SFY LPS+++GN + +KAAYS+YNV
Sbjct 240 ENDVEHWLLPREDVVDSYLVESPETHDVTDFCSFYTLPSTILGNPNYTTLKAAYSYYNVA 299
Query 60 TSAPLKELIQDALCLAKQKDFDVFNALDVMENKTFVEDLKFGIGDGFLRYYLYNWRCPQV 119
T +L+ DAL ++KQK FDVFNALDVM N++F+++LKFG GDG L YYLYN+R
Sbjct 300 TQTSFLQLMNDALIVSKQKGFDVFNALDVMHNESFLKELKFGPGDGQLHYYLYNYRLKSA 359
Query 120 LHN-DIGLVLL 129
L ++GLVLL
Sbjct 360 LKPAELGLVLL 370
> CE01406
Length=450
Score = 135 bits (339), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 71/129 (55%), Positives = 94/129 (72%), Gaps = 1/129 (0%)
Query 1 TQEEVRHWLMPKEGVVHVYVRE-QGGKVTDLISFYELPSSVIGNQKHKEVKAAYSFYNVG 59
++EE+ H L+PK+GVV+ YV E Q GK+TD +SFY LPS+V+G+ HK + AAY +Y V
Sbjct 321 SEEELAHALVPKKGVVYSYVAENQNGKITDFVSFYSLPSTVMGHTTHKTIYAAYLYYYVA 380
Query 60 TSAPLKELIQDALCLAKQKDFDVFNALDVMENKTFVEDLKFGIGDGFLRYYLYNWRCPQV 119
S K+LI D+L LA ++ FDVFNALD+M N+ DLKFG GDG L+YYLYNW+C +
Sbjct 381 GSVTPKQLINDSLILANREKFDVFNALDLMHNEKIFSDLKFGKGDGNLQYYLYNWKCADM 440
Query 120 LHNDIGLVL 128
+ IGLVL
Sbjct 441 KPSQIGLVL 449
> CE28669
Length=452
Score = 135 bits (339), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 71/129 (55%), Positives = 94/129 (72%), Gaps = 1/129 (0%)
Query 1 TQEEVRHWLMPKEGVVHVYVRE-QGGKVTDLISFYELPSSVIGNQKHKEVKAAYSFYNVG 59
++EE+ H L+PK+GVV+ YV E Q GK+TD +SFY LPS+V+G+ HK + AAY +Y V
Sbjct 323 SEEELAHALVPKKGVVYSYVAENQNGKITDFVSFYSLPSTVMGHTTHKTIYAAYLYYYVA 382
Query 60 TSAPLKELIQDALCLAKQKDFDVFNALDVMENKTFVEDLKFGIGDGFLRYYLYNWRCPQV 119
S K+LI D+L LA ++ FDVFNALD+M N+ DLKFG GDG L+YYLYNW+C +
Sbjct 383 GSVTPKQLINDSLILANREKFDVFNALDLMHNEKIFSDLKFGKGDGNLQYYLYNWKCADM 442
Query 120 LHNDIGLVL 128
+ IGLVL
Sbjct 443 KPSQIGLVL 451
> SPBC2G2.11
Length=466
Score = 121 bits (304), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 64/140 (45%), Positives = 88/140 (62%), Gaps = 21/140 (15%)
Query 1 TQEEVRHWLMPKE-----GVVHVYVRE--QGGKVTDLISFYELPSSVIGNQKHKEVKAAY 53
++EEVRHW + + VV YV E + K+TD SFY LPS+VIGN K+K+++AAY
Sbjct 304 SEEEVRHWFLYTDKVSSGPVVWSYVVENPESKKITDFFSFYSLPSTVIGNPKYKDIQAAY 363
Query 54 SFYNVGTSAP--------------LKELIQDALCLAKQKDFDVFNALDVMENKTFVEDLK 99
+Y S P K ++ DAL LAK+ FDVFNA+ V++N F++DLK
Sbjct 364 LYYYASDSCPKDLSSESQLAFVERCKLIVNDALILAKKFHFDVFNAVTVLDNNLFLKDLK 423
Query 100 FGIGDGFLRYYLYNWRCPQV 119
FG GDGFL YY+YN+ CP++
Sbjct 424 FGEGDGFLNYYIYNYNCPKI 443
> At2g44170
Length=389
Score = 100 bits (248), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 52/128 (40%), Positives = 82/128 (64%), Gaps = 3/128 (2%)
Query 4 EVRHWLMPKEGVVHVYV--REQGGKVTDLISFYELPSSVIGNQKHKEVKAAYSFYNVGTS 61
+V+HWL+P+E +V+ YV + VTD SF ++ GN+K+ ++ AY+ NV T
Sbjct 262 DVKHWLLPRENIVYSYVVVSPETHDVTDFCSFCNSSITIPGNRKYTTLECAYACCNVATL 321
Query 62 APLKELIQDALCLAKQKDFDVFNALDVMENKTFVEDLKFGIGDGFLRYYLYNWRCPQVLH 121
L +L+ DAL ++KQK FDVF A DVM+N++F+++L+F YYLYN+R L
Sbjct 322 TSLSQLVNDALIVSKQKGFDVFYASDVMQNESFLKELRFYPLCRQSHYYLYNYRLRNALK 381
Query 122 -NDIGLVL 128
+++GL+L
Sbjct 382 PSELGLIL 389
> YLR195c
Length=455
Score = 91.7 bits (226), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 56/143 (39%), Positives = 78/143 (54%), Gaps = 24/143 (16%)
Query 1 TQEEVRHWLMPKEG------VVHVYVREQ-GGKVTDLISFYELPSSVIGNQKHKEVKAAY 53
T+EE H + +E V+ YV EQ GK+TD SFY LP +++ N K+K++ Y
Sbjct 290 TKEEFEHNFIGEESLPLDKQVIFSYVVEQPDGKITDFFSFYSLPFTILNNTKYKDLGIGY 349
Query 54 SFY-------------NVGTSAPLK----ELIQDALCLAKQKDFDVFNALDVMENKTFVE 96
+Y + + LK ELI DA LAK + DVFNAL +N F++
Sbjct 350 LYYYATDADFQFKDRFDPKATKALKTRLCELIYDACILAKNANMDVFNALTSQDNTLFLD 409
Query 97 DLKFGIGDGFLRYYLYNWRCPQV 119
DLKFG GDGFL +YL+N+R +
Sbjct 410 DLKFGPGDGFLNFYLFNYRAKPI 432
> ECU03g1350
Length=355
Score = 73.9 bits (180), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 38/126 (30%), Positives = 69/126 (54%), Gaps = 4/126 (3%)
Query 3 EEVRHWLMPKEGVVHVYVREQGGKVTDLISFYELPSSVIGNQKHKEVKAAYSFYNVGTSA 62
E++ P + VV+ YV E G + +F+ + + I + ++++ Y +Y G
Sbjct 233 EDIVSTFRPVKNVVYTYVHESNGAIDGFGAFFVVGT--IEKKSGRQIQGGYLYYRGGKD- 289
Query 63 PLKELIQDALCLAKQKDFDVFNALDVMENKTFVEDLKFGIGDGFLRYYLYNWRCPQVLHN 122
+ E++ D + ++ + DVFN LD+MEN +F+ L F G G +RYYLYNW+ ++ +
Sbjct 290 -VAEMVGDLMHFSQVEGCDVFNCLDMMENSSFLARLGFVCGSGEIRYYLYNWKSEEIPRD 348
Query 123 DIGLVL 128
+ VL
Sbjct 349 KVFFVL 354
> YLR197w
Length=504
Score = 30.8 bits (68), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 33/63 (52%), Gaps = 1/63 (1%)
Query 13 EGVVHVYVREQGGKVTDLISFYELPSSVIGNQKHKEVKAAYSFYNVGTSAPLKEL-IQDA 71
+G + Y+ + + + ++ E PS+V G+ K+V+ FYN G EL IQ+A
Sbjct 372 KGRISRYLANKCSMASRIDNYSEEPSNVFGSVLKKQVEQRLEFYNTGKPTLKNELAIQEA 431
Query 72 LCL 74
+ L
Sbjct 432 MEL 434
> At5g40480
Length=1919
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 26/58 (44%), Gaps = 10/58 (17%)
Query 54 SFYNVGTSAPLKELIQDALCLAKQKDFDVF-----NALDVMENKTFVEDLKFGIGDGF 106
+ Y++G S PL AL L K D D + + +ME T DL GI DG
Sbjct 1014 TVYDIGVSPPLS-----ALALIKVADVDWIKIASGDEISIMEGSTHSIDLLTGIDDGM 1066
> Hs5453794_1
Length=498
Score = 29.3 bits (64), Expect = 2.1, Method: Composition-based stats.
Identities = 16/55 (29%), Positives = 28/55 (50%), Gaps = 1/55 (1%)
Query 13 EGVVHVYVREQGGKVTDLISFYELPSSVIGNQKHKEVKAAYSFYNVGTSAPLKEL 67
+G + Y+ + + + F E+P+SV G + ++V+ SFY G P K L
Sbjct 371 KGRISRYLANKCSIASRIDCFSEVPTSVFGEKLREQVEERLSFYETG-EIPRKNL 424
> At4g37870
Length=671
Score = 28.9 bits (63), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 27/111 (24%), Positives = 49/111 (44%), Gaps = 22/111 (19%)
Query 4 EVRHWLMPKEGVVHVYVREQGGKVTDLISFYELPSS------------VIGNQKHKEVKA 51
V H+LMPK ++ ++ GK D+ F+ L + +IG+ +H +
Sbjct 338 SVMHYLMPKRRILSLHSGCNMGKDGDVALFFGLSGTGKTTLSTDHNRYLIGDDEHCWTET 397
Query 52 AYSFYNVGTSAPLKELIQDALCLAKQKDFDVFNAL---DVMENKTFVEDLK 99
S G A + L+++K+ D++NA+ V+EN F E +
Sbjct 398 GVSNIEGGCYA-------KCVDLSREKEPDIWNAIKFGTVLENVVFDEHTR 441
> Hs13384594
Length=697
Score = 28.5 bits (62), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 27/49 (55%), Gaps = 0/49 (0%)
Query 66 ELIQDALCLAKQKDFDVFNALDVMENKTFVEDLKFGIGDGFLRYYLYNW 114
+LI D L KQK V L+ + N+TFV +LK G + L +++W
Sbjct 199 KLIPDPRNLTKQKTRTVKATLNPVWNETFVFNLKPGDVERRLSVEVWDW 247
> At1g72740
Length=289
Score = 28.1 bits (61), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 19/60 (31%), Positives = 30/60 (50%), Gaps = 3/60 (5%)
Query 32 SFYELPS---SVIGNQKHKEVKAAYSFYNVGTSAPLKELIQDALCLAKQKDFDVFNALDV 88
+FY++P + IG K KE N TSA +++I++A A K + N +DV
Sbjct 187 NFYKIPDPSGTKIGVPKPKETHTKLRQANNQTSADSQQMIEEAAITAACKVVEAENKIDV 246
> At1g61550
Length=802
Score = 27.7 bits (60), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 11/32 (34%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 10 MPKEGVVHVYVREQGGKVTDLISFYELPSSVI 41
+PKE V+ + G + +DLI+ E+ SV+
Sbjct 768 LPKEPTFAVHTSDDGSRTSDLITVNEVTQSVV 799
Lambda K H
0.322 0.140 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1209785478
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40