bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2022_orf1
Length=148
Score E
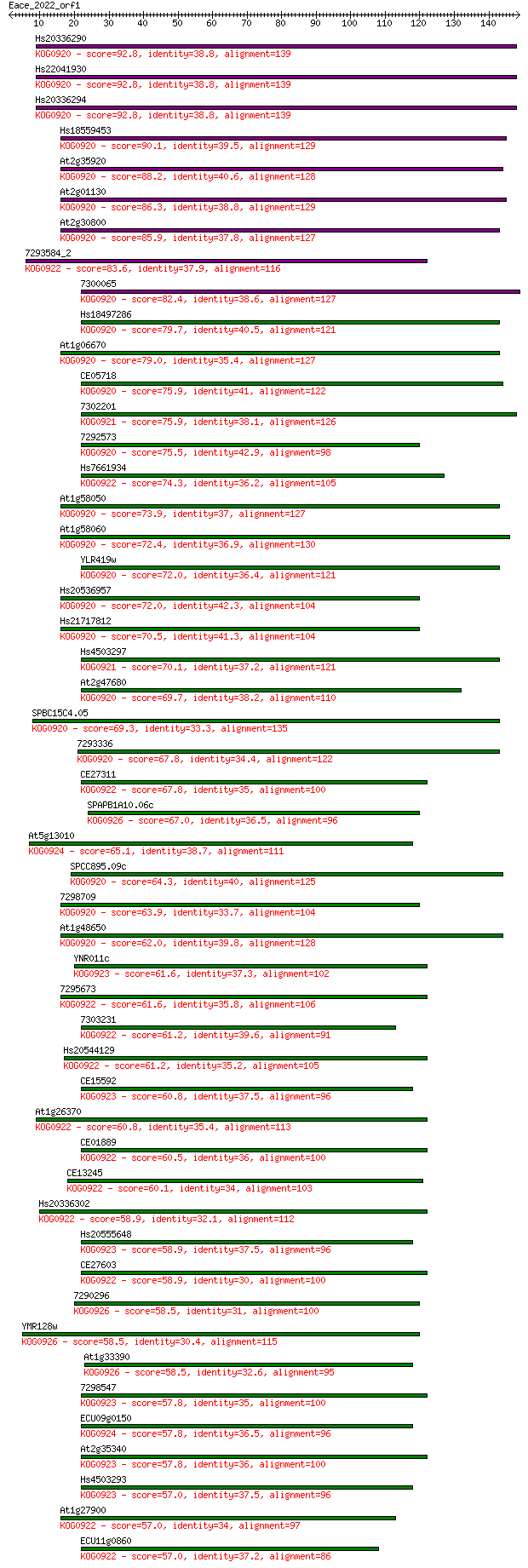
Sequences producing significant alignments: (Bits) Value
Hs20336290 92.8 2e-19
Hs22041930 92.8 2e-19
Hs20336294 92.8 2e-19
Hs18559453 90.1 1e-18
At2g35920 88.2 5e-18
At2g01130 86.3 2e-17
At2g30800 85.9 2e-17
7293584_2 83.6 1e-16
7300065 82.4 3e-16
Hs18497286 79.7 2e-15
At1g06670 79.0 3e-15
CE05718 75.9 3e-14
7302201 75.9 3e-14
7292573 75.5 3e-14
Hs7661934 74.3 7e-14
At1g58050 73.9 1e-13
At1g58060 72.4 3e-13
YLR419w 72.0 3e-13
Hs20536957 72.0 4e-13
Hs21717812 70.5 1e-12
Hs4503297 70.1 2e-12
At2g47680 69.7 2e-12
SPBC15C4.05 69.3 2e-12
7293336 67.8 6e-12
CE27311 67.8 7e-12
SPAPB1A10.06c 67.0 1e-11
At5g13010 65.1 5e-11
SPCC895.09c 64.3 8e-11
7298709 63.9 1e-10
At1g48650 62.0 4e-10
YNR011c 61.6 5e-10
7295673 61.6 5e-10
7303231 61.2 6e-10
Hs20544129 61.2 7e-10
CE15592 60.8 8e-10
At1g26370 60.8 8e-10
CE01889 60.5 1e-09
CE13245 60.1 1e-09
Hs20336302 58.9 3e-09
Hs20555648 58.9 3e-09
CE27603 58.9 4e-09
7290296 58.5 4e-09
YMR128w 58.5 4e-09
At1g33390 58.5 4e-09
7298547 57.8 8e-09
ECU09g0150 57.8 8e-09
At2g35340 57.8 8e-09
Hs4503293 57.0 1e-08
At1g27900 57.0 1e-08
ECU11g0860 57.0 1e-08
> Hs20336290
Length=1155
Score = 92.8 bits (229), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 54/139 (38%), Positives = 79/139 (56%), Gaps = 3/139 (2%)
Query 9 KRAKQHAPSSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWAS 68
++A P KI+LATNIAE+SIT D+ V+D L KE+ TK L+ W S
Sbjct 674 QKAIFQQPPVGVRKIVLATNIAETSITINDIVHVVDSGLHKEERYDLKTKVSCLETVWVS 733
Query 69 RAALDQRRGRTGRVRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMAALPGEST 128
RA + QRRGR GR + G L PR + PF +PE+ R PLE++++ +P ++
Sbjct 734 RANVIQRRGRAGRCQSGFAYHLFPRSRLEKMVPFQVPEILRTPLENLVLQAKIHMPEKT- 792
Query 129 DASLFFLSQAQDPPDVGRV 147
++ FLS+A D P++ V
Sbjct 793 --AVEFLSKAVDSPNIKAV 809
> Hs22041930
Length=1035
Score = 92.8 bits (229), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 54/139 (38%), Positives = 79/139 (56%), Gaps = 3/139 (2%)
Query 9 KRAKQHAPSSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWAS 68
++A P KI+LATNIAE+SIT D+ V+D L KE+ TK L+ W S
Sbjct 554 QKAIFQQPPVGVRKIVLATNIAETSITINDIVHVVDSGLHKEERYDLKTKVSCLETVWVS 613
Query 69 RAALDQRRGRTGRVRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMAALPGEST 128
RA + QRRGR GR + G L PR + PF +PE+ R PLE++++ +P ++
Sbjct 614 RANVIQRRGRAGRCQSGFAYHLFPRSRLEKMVPFQVPEILRTPLENLVLQAKIHMPEKT- 672
Query 129 DASLFFLSQAQDPPDVGRV 147
++ FLS+A D P++ V
Sbjct 673 --AVEFLSKAVDSPNIKAV 689
> Hs20336294
Length=1194
Score = 92.8 bits (229), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 54/139 (38%), Positives = 79/139 (56%), Gaps = 3/139 (2%)
Query 9 KRAKQHAPSSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWAS 68
++A P KI+LATNIAE+SIT D+ V+D L KE+ TK L+ W S
Sbjct 713 QKAIFQQPPVGVRKIVLATNIAETSITINDIVHVVDSGLHKEERYDLKTKVSCLETVWVS 772
Query 69 RAALDQRRGRTGRVRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMAALPGEST 128
RA + QRRGR GR + G L PR + PF +PE+ R PLE++++ +P ++
Sbjct 773 RANVIQRRGRAGRCQSGFAYHLFPRSRLEKMVPFQVPEILRTPLENLVLQAKIHMPEKT- 831
Query 129 DASLFFLSQAQDPPDVGRV 147
++ FLS+A D P++ V
Sbjct 832 --AVEFLSKAVDSPNIKAV 848
> Hs18559453
Length=965
Score = 90.1 bits (222), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 51/129 (39%), Positives = 71/129 (55%), Gaps = 4/129 (3%)
Query 16 PSSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQR 75
P KI+LATNIAE+ IT DV V+D KE H ++ L + S+A+ QR
Sbjct 515 PPPGVRKIVLATNIAETGITIPDVVFVIDTGRTKENKYHESSQMSSLVETFVSKASALQR 574
Query 76 RGRTGRVRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMAALPGESTDASLFFL 135
+GR GRVRDG + RE + + +PE+ R+PLE++ + IM G D FL
Sbjct 575 QGRAGRVRDGFCFRMYTRERFEGFMDYSVPEILRVPLEELCLHIMKCNLGSPED----FL 630
Query 136 SQAQDPPDV 144
S+A DPP +
Sbjct 631 SKALDPPQL 639
> At2g35920
Length=993
Score = 88.2 bits (217), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 52/128 (40%), Positives = 71/128 (55%), Gaps = 4/128 (3%)
Query 16 PSSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQR 75
P KI+LATNIAESSIT DV V+D KE A+ K L W S+A+ QR
Sbjct 554 PPPNKRKIVLATNIAESSITIDDVVYVVDCGKAKETSYDALNKVACLLPSWISKASAHQR 613
Query 76 RGRTGRVRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMAALPGESTDASLFFL 135
RGR GRV+ G L P+ +Y++ + LPE+ R PL+++ + I + G FL
Sbjct 614 RGRAGRVQAGVCYRLYPKVIYDAFPQYQLPEIIRTPLQELCLHIKSLQVGSIGS----FL 669
Query 136 SQAQDPPD 143
++A PPD
Sbjct 670 AKALQPPD 677
> At2g01130
Length=1112
Score = 86.3 bits (212), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 50/129 (38%), Positives = 71/129 (55%), Gaps = 4/129 (3%)
Query 16 PSSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQR 75
P+S KI+LATNIAE+SIT DV V+D KE A+ L W S+ + QR
Sbjct 550 PASGVRKIVLATNIAETSITINDVAFVIDCGKAKETSYDALNNTPCLLPSWISKVSAQQR 609
Query 76 RGRTGRVRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMAALPGESTDASLFFL 135
RGR GRVR G L P+ +Y++ + LPE+ R PL + + I + G ++ FL
Sbjct 610 RGRAGRVRPGQCYHLYPKCVYDAFAEYQLPEILRTPLHSLCLQIKSLNLGSISE----FL 665
Query 136 SQAQDPPDV 144
S+A P++
Sbjct 666 SRALQSPEL 674
> At2g30800
Length=1299
Score = 85.9 bits (211), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 48/127 (37%), Positives = 68/127 (53%), Gaps = 3/127 (2%)
Query 16 PSSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQR 75
P C KI+LATNIAES++T DV V+D +KEK LQ W S+A QR
Sbjct 604 PPPGCRKIVLATNIAESAVTIDDVVYVIDSGRMKEKSYDPYNNVSTLQSSWVSKANAKQR 663
Query 76 RGRTGRVRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMAALPGESTDASLFFL 135
+GR GR + G L R S+ F +PE++R+P+E++ + + P T+ FL
Sbjct 664 QGRAGRCQPGICYHLYSRLRAASMPDFKVPEIKRMPVEELCLQVKILDPNCKTND---FL 720
Query 136 SQAQDPP 142
+ DPP
Sbjct 721 QKLLDPP 727
> 7293584_2
Length=838
Score = 83.6 bits (205), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 44/116 (37%), Positives = 67/116 (57%), Gaps = 1/116 (0%)
Query 6 GDLKRAKQHAPSSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLR 65
D + +AP K I++TNIAE+S+T V V+D VKE + A K RL+
Sbjct 285 ADQSKVFDYAPEGM-RKCIVSTNIAETSLTVDGVRFVVDSGKVKEMNFDATCKGQRLKEF 343
Query 66 WASRAALDQRRGRTGRVRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMA 121
W S+++ DQR+GR GR G L E YN+ E +P PE+ R+PL+ +L+ +++
Sbjct 344 WVSKSSADQRKGRAGRTGPGVCFRLYTAEQYNAFEAYPTPEIYRVPLDTMLLQMVS 399
> 7300065
Length=1434
Score = 82.4 bits (202), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 49/128 (38%), Positives = 69/128 (53%), Gaps = 5/128 (3%)
Query 22 KIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGR 81
KIIL TNIAESSIT DV V+DF L K K + L+L WAS+A QR GR GR
Sbjct 424 KIILTTNIAESSITVPDVSYVIDFCLAKVKVTDTASSFSSLRLTWASKANCRQRAGRVGR 483
Query 82 VRDGTYLCLIPRELYNSLEP-FPLPELQRLPLEDVLVDIMAALPGESTDASLFFLSQAQD 140
+R G ++ + Y P F +PE+ RLPL++ ++ G + L+ A
Sbjct 484 LRSGRVYRMVNKHFYQREMPEFGIPEMLRLPLQNSVLKAKVLNMGSPVE----ILALALS 539
Query 141 PPDVGRVY 148
PP++ ++
Sbjct 540 PPNLSDIH 547
> Hs18497286
Length=1008
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 49/126 (38%), Positives = 66/126 (52%), Gaps = 14/126 (11%)
Query 22 KIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGR 81
KI++ATNIAE+SIT DV V+D +KE H + W S+A QR+GR GR
Sbjct 546 KIVIATNIAETSITIDDVVYVIDGGKIKETHFDTQNNISTMSAEWVSKANAKQRKGRAGR 605
Query 82 VRDGTYLCLIPRELYNS-----LEPFPLPELQRLPLEDVLVDIMAALPGESTDASLFFLS 136
V+ G C LYN L+ + LPE+ R PLE++ + I G +FLS
Sbjct 606 VQPGH--CY---HLYNGLRASLLDDYQLPEILRTPLEELCLQIKILRLG----GIAYFLS 656
Query 137 QAQDPP 142
+ DPP
Sbjct 657 RLMDPP 662
> At1g06670
Length=1576
Score = 79.0 bits (193), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 45/127 (35%), Positives = 66/127 (51%), Gaps = 3/127 (2%)
Query 16 PSSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQR 75
P C KI+LATNIAES++T DV V+D +KEK LQ W S+A QR
Sbjct 628 PPRGCRKIVLATNIAESAVTIDDVVYVIDSGRMKEKSYDPYNDVSTLQSSWVSKANAKQR 687
Query 76 RGRTGRVRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMAALPGESTDASLFFL 135
GR GR + G L + SL + +PE+ R+P++++ + + P + + FL
Sbjct 688 AGRAGRCQAGICYHLYSKLRAASLPEYRVPEVMRMPVDELCLQVKMLDPNCNVND---FL 744
Query 136 SQAQDPP 142
+ DPP
Sbjct 745 QKLMDPP 751
> CE05718
Length=1425
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 50/122 (40%), Positives = 67/122 (54%), Gaps = 2/122 (1%)
Query 22 KIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGR 81
KIILATNIAE+SIT DV V+D VKEK K L ++ +R+ DQR GR GR
Sbjct 704 KIILATNIAEASITIEDVIFVVDTGKVKEKSFDHEAKLSTLTVKPIARSNADQRSGRAGR 763
Query 82 VRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMAALPGESTDASLFFLSQAQDP 141
V +G + L + YNS+ + E++R + DV + A L T FLS A +P
Sbjct 764 VANGYCIRLYTEQEYNSMPETQIAEMKRAAIYDVT--LHAKLFAPKTLKISEFLSLAPEP 821
Query 142 PD 143
P+
Sbjct 822 PE 823
> 7302201
Length=1293
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 48/126 (38%), Positives = 66/126 (52%), Gaps = 4/126 (3%)
Query 22 KIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGR 81
KIIL+TNIAE+SIT D+ V+D + K + WAS+ L+QR+GR GR
Sbjct 712 KIILSTNIAETSITIDDIVFVIDICKARMKLFTSHNNLTSYATVWASKTNLEQRKGRAGR 771
Query 82 VRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMAALPGESTDASLFFLSQAQDP 141
VR G L R + +LE PE+ R PL ++ + I G + FLS+A +P
Sbjct 772 VRPGFCFTLCSRARFQALEDNLTPEMFRTPLHEMALTIKLLRLG----SIHHFLSKALEP 827
Query 142 PDVGRV 147
P V V
Sbjct 828 PPVDAV 833
> 7292573
Length=1280
Score = 75.5 bits (184), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 42/99 (42%), Positives = 58/99 (58%), Gaps = 1/99 (1%)
Query 22 KIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGR 81
KI+L+TNIAE+S+T D V+D L+KEK + L L W SRA QR+GR GR
Sbjct 804 KIVLSTNIAETSVTIDDCVFVVDCGLMKEKCFDSNRNMESLDLVWVSRANAKQRKGRAGR 863
Query 82 VRDGTYLCLIPRELYN-SLEPFPLPELQRLPLEDVLVDI 119
V G + L Y + P+PE+QR+PLE +++ I
Sbjct 864 VMPGVCIHLYTSYRYQYHILAQPVPEIQRVPLEQIVLRI 902
> Hs7661934
Length=576
Score = 74.3 bits (181), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 38/105 (36%), Positives = 61/105 (58%), Gaps = 0/105 (0%)
Query 22 KIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGR 81
K IL+TNIAE+S+T + V+D VKE K RLQ W S+A+ +QR+GR GR
Sbjct 350 KCILSTNIAETSVTIDGIRFVVDSGKVKEMSYDPQAKLQRLQEFWISQASAEQRKGRAGR 409
Query 82 VRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMAALPGE 126
G L Y++ P+P+PE++R+ L+ +++ + + G+
Sbjct 410 TGPGVCFRLYAESDYDAFAPYPVPEIRRVALDSLVLQMKSMSVGD 454
> At1g58050
Length=1417
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 47/128 (36%), Positives = 66/128 (51%), Gaps = 5/128 (3%)
Query 16 PSSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQR 75
P K+I+ATNIAE+SIT DV V+D KE + K + W S+A QR
Sbjct 935 PPKGIRKVIIATNIAETSITIEDVVYVIDSGKHKENRYNPHKKLSSMVEDWVSKANARQR 994
Query 76 RGRTGRVRDGTYLCLIPRELYNSL-EPFPLPELQRLPLEDVLVDIMAALPGESTDASLFF 134
GR GRV+ G L R + L P+ +PE+ R+PL ++ + I G+ F
Sbjct 995 MGRAGRVKPGHCFSLYTRHRFEKLMRPYQVPEMLRVPLVELCLHIKLLGLGQIKP----F 1050
Query 135 LSQAQDPP 142
LS+A +PP
Sbjct 1051 LSKALEPP 1058
> At1g58060
Length=1453
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 48/131 (36%), Positives = 65/131 (49%), Gaps = 5/131 (3%)
Query 16 PSSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQR 75
P K+I ATNIAE+SIT DV V+D KE + K + W S+A QR
Sbjct 968 PPKGLRKVIAATNIAETSITIDDVVYVIDSGKHKENRYNPQKKLSSMVEDWISQANARQR 1027
Query 76 RGRTGRVRDGTYLCLIPRELYNSL-EPFPLPELQRLPLEDVLVDIMAALPGESTDASLFF 134
GR GRV+ G L R + L P+ +PE+ R+PL ++ + I G F
Sbjct 1028 TGRAGRVKPGICFSLYTRYRFEKLMRPYQVPEMLRMPLVELCLQIKLLGLGHIKP----F 1083
Query 135 LSQAQDPPDVG 145
LS+A +PP G
Sbjct 1084 LSRALEPPSEG 1094
> YLR419w
Length=1435
Score = 72.0 bits (175), Expect = 3e-13, Method: Composition-based stats.
Identities = 44/122 (36%), Positives = 67/122 (54%), Gaps = 5/122 (4%)
Query 22 KIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGR 81
K++++TNIAE+SIT D +D K + +L + S+A + QRRGR GR
Sbjct 918 KVVVSTNIAETSITIDDCVATIDTGRAKSMFYNPKDNTTKLIESFISKAEVKQRRGRAGR 977
Query 82 VRDGTYLCLIPRELY-NSLEPFPLPELQRLPLEDVLVDIMAALPGESTDASLFFLSQAQD 140
VR+G L + LY N + P+PE++R+PLE + + + A+ + A FLS A D
Sbjct 978 VREGLSYKLFSKNLYENDMISMPIPEIKRIPLESLYLSV-KAMGIKDVKA---FLSTALD 1033
Query 141 PP 142
P
Sbjct 1034 AP 1035
> Hs20536957
Length=688
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 44/105 (41%), Positives = 58/105 (55%), Gaps = 1/105 (0%)
Query 16 PSSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQR 75
P + KII++TNIAE+SIT DV V+D +KEK A L+ + S+A QR
Sbjct 204 PPAGVTKIIISTNIAETSITIDDVVYVIDSGKMKEKRYDASKGMESLEDTFVSQANALQR 263
Query 76 RGRTGRVRDGTYLCLIPRELYN-SLEPFPLPELQRLPLEDVLVDI 119
+GR GRV G L YN L LPE+QR+PLE + + I
Sbjct 264 KGRAGRVASGVCFHLFTSHHYNHQLLKQQLPEIQRVPLEQLCLRI 308
> Hs21717812
Length=860
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 43/105 (40%), Positives = 58/105 (55%), Gaps = 1/105 (0%)
Query 16 PSSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQR 75
P + KII++TNIAE+SIT DV V+D +KEK A L+ + S+A QR
Sbjct 376 PPAGVTKIIISTNIAETSITIDDVVYVIDSGKMKEKRYDASKGMESLEDTFVSQANALQR 435
Query 76 RGRTGRVRDGTYLCLIPRELYN-SLEPFPLPELQRLPLEDVLVDI 119
+GR GRV G + YN L LPE+QR+PLE + + I
Sbjct 436 KGRAGRVASGVCFHVFTSHHYNHQLLKQQLPEIQRVPLEQLCLRI 480
> Hs4503297
Length=1279
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 45/121 (37%), Positives = 60/121 (49%), Gaps = 4/121 (3%)
Query 22 KIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGR 81
K+IL+TNIAE+SIT DV V+D K K A WAS+ L+QR+GR GR
Sbjct 707 KVILSTNIAETSITINDVVYVIDSCKQKVKLFTAHNNMTNYSTVWASKTNLEQRKGRAGR 766
Query 82 VRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMAALPGESTDASLFFLSQAQDP 141
G L R + LE PE+ R PL ++ + I G FL++A +P
Sbjct 767 STAGFCFHLCSRARFERLETHMTPEMFRTPLHEIALSIKLLRLGGIGQ----FLAKAIEP 822
Query 142 P 142
P
Sbjct 823 P 823
> At2g47680
Length=1015
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 42/110 (38%), Positives = 56/110 (50%), Gaps = 0/110 (0%)
Query 22 KIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGR 81
K+ILATNIAESS+T V V+D + K +QL W SR+ +QRRGRTGR
Sbjct 323 KVILATNIAESSVTIPKVAYVIDSCRSLQVFWDPSRKRDAVQLVWVSRSQAEQRRGRTGR 382
Query 82 VRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMAALPGESTDAS 131
DG L+P +N LE P + +L L ++ I DA+
Sbjct 383 TCDGEVYRLVPSAFFNKLEEHEPPSILKLSLRQQVLHICCTESRAINDAN 432
> SPBC15C4.05
Length=1428
Score = 69.3 bits (168), Expect = 2e-12, Method: Composition-based stats.
Identities = 45/140 (32%), Positives = 73/140 (52%), Gaps = 9/140 (6%)
Query 8 LKRAKQHA----PSSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQ 63
L A+Q + P C KI+L+TNIAE+ +T DV V+D + +E ++ RL
Sbjct 944 LSSAQQQSVFNIPPKGCRKIVLSTNIAETGVTIPDVTCVIDTGVHREMRYNSRRHLSRLT 1003
Query 64 LRWASRAALDQRRGRTGRVRDGTYLCLIPRELYNS-LEPFPLPELQRLPLEDVLVDIMAA 122
+ S+A QR GR GRV++G L + +++ + PE+ RL L++V++ +
Sbjct 1004 DTFVSKANAKQRSGRAGRVQEGICYHLFSKFKHDTQFLSYQTPEILRLNLQEVVLRVKMC 1063
Query 123 LPGESTDASLFFLSQAQDPP 142
G+ D L +A DPP
Sbjct 1064 QMGDVQDV----LGKALDPP 1079
> 7293336
Length=976
Score = 67.8 bits (164), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 42/122 (34%), Positives = 63/122 (51%), Gaps = 3/122 (2%)
Query 21 MKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTG 80
+KIIL+TNI ++SIT D+ V+D L K K + A +L L W S+A QR GR G
Sbjct 515 LKIILSTNIGQTSITIPDLLYVIDTGLAKMKTYDSTIDASQLTLTWISQADAKQRAGRAG 574
Query 81 RVRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMAALPGESTDASLFFLSQAQD 140
RV G L + + + +PE+ R L+++ + A P + + FL+ A D
Sbjct 575 RVCHGNCYRLYDNDRMARMNLYTIPEIMRRTLDEICLLTKLAAPDKKIEN---FLALALD 631
Query 141 PP 142
P
Sbjct 632 TP 633
> CE27311
Length=732
Score = 67.8 bits (164), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 53/100 (53%), Gaps = 0/100 (0%)
Query 22 KIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGR 81
K+++ATNIAE+SIT +C V+D VK + HA L S+A+ +QR GR GR
Sbjct 356 KVVVATNIAEASITIPGICYVIDTGYVKLRAQHAANGVETLMRVTVSKASAEQRAGRAGR 415
Query 82 VRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMA 121
+R G L P + +PE+QR + ++ + A
Sbjct 416 IRPGKCYRLYPESEFERFAEGTVPEIQRCQMASTILQLKA 455
> SPAPB1A10.06c
Length=1183
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 35/97 (36%), Positives = 56/97 (57%), Gaps = 1/97 (1%)
Query 24 ILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGRVR 83
I+ATN+AE+SIT ++ V+D KE+ + T + ++RW S+A DQR GR GR
Sbjct 731 IVATNVAETSITIPNIRYVVDCGKAKERVYNEKTSVQKFEVRWISKANADQRAGRAGRTG 790
Query 84 DGTYLCLIPRELYNSLEPF-PLPELQRLPLEDVLVDI 119
G L +++S P LPE+ R P+E +++ +
Sbjct 791 PGHCYRLYSSAVFDSSFPLHSLPEILRTPVESIVLQM 827
> At5g13010
Length=1226
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 43/112 (38%), Positives = 57/112 (50%), Gaps = 1/112 (0%)
Query 7 DLKRAKQHAPSSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRW 66
DL+ P K I+ATNIAE+S+T + V+D K K + LQ+
Sbjct 787 DLQAKIFQKPEDGARKCIVATNIAETSLTVDGIYYVIDTGYGKMKVFNPRMGMDALQVFP 846
Query 67 ASRAALDQRRGRTGRVRDGTYLCLIPRELY-NSLEPFPLPELQRLPLEDVLV 117
SRAA DQR GR GR GT L Y N + P P+PE+QR L +V++
Sbjct 847 ISRAASDQRAGRAGRTGPGTCYRLYTESAYLNEMLPSPVPEIQRTNLGNVVL 898
> SPCC895.09c
Length=1327
Score = 64.3 bits (155), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 50/134 (37%), Positives = 65/134 (48%), Gaps = 10/134 (7%)
Query 19 TCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGR 78
T KII ATNIAE+SIT DV V+D VK+ + WASRAA QRRGR
Sbjct 863 TKRKIICATNIAETSITIDDVVAVIDSGRVKQIDYDVERDLVTFKETWASRAACQQRRGR 922
Query 79 TGRVRDGTYLCLIPR--ELYNSLEPFPLPELQRLPLEDVLVDIMAALP-----GESTDAS 131
GRV+ G L R E L P PE+ R L V ++++ + G S +
Sbjct 923 AGRVKKGICYKLYTRGFEEKGMLGQTP-PEVLRTALSQVCLNVVPLVKRFSSAGNSVNQG 981
Query 132 LF--FLSQAQDPPD 143
F++ DPP+
Sbjct 982 SIKKFMNSLIDPPN 995
> 7298709
Length=674
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 35/104 (33%), Positives = 54/104 (51%), Gaps = 0/104 (0%)
Query 16 PSSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQR 75
P + K+I++T IAE+S+T DV V++ K + T L W ++A QR
Sbjct 407 PPAGQRKVIISTIIAETSVTIDDVVYVINSGRTKATNYDIETNIQSLDEVWVTKANTQQR 466
Query 76 RGRTGRVRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDI 119
RGR GRVR G L R + ++ P PE+ R LE +++ +
Sbjct 467 RGRAGRVRPGICYNLFSRAREDRMDDIPTPEILRSKLESIILSL 510
> At1g48650
Length=1167
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 51/128 (39%), Positives = 68/128 (53%), Gaps = 4/128 (3%)
Query 16 PSSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQR 75
P KI+LATN+AE+SIT DV V+D KE A+ L W S+AA QR
Sbjct 601 PPEGIRKIVLATNMAETSITINDVVYVIDCGKAKETSYDALNNTPCLLPSWISKAAARQR 660
Query 76 RGRTGRVRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMAALPGESTDASLFFL 135
RGR GRV G L PR +Y + + PEL R PL+ + + I + G ++ FL
Sbjct 661 RGRAGRVMPGECYHLYPRCVYEAFADYQQPELLRTPLQSLCLQIKSLGLGSISE----FL 716
Query 136 SQAQDPPD 143
S+A PP+
Sbjct 717 SRALQPPE 724
> YNR011c
Length=876
Score = 61.6 bits (148), Expect = 5e-10, Method: Composition-based stats.
Identities = 38/103 (36%), Positives = 57/103 (55%), Gaps = 1/103 (0%)
Query 20 CMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRT 79
C K++LATNIAE+S+T + V+D VKE T +L SRA++DQR GR
Sbjct 494 CRKVVLATNIAETSLTIDGIRYVIDPGFVKENSYVPSTGMTQLLTVPCSRASVDQRAGRA 553
Query 80 GRVRDGTYLCLIPRELY-NSLEPFPLPELQRLPLEDVLVDIMA 121
GRV G + + Y + LE P PE+ R L + ++ +++
Sbjct 554 GRVGPGKCFRIFTKWSYLHELELMPKPEITRTNLSNTVLLLLS 596
> 7295673
Length=671
Score = 61.6 bits (148), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 38/106 (35%), Positives = 56/106 (52%), Gaps = 0/106 (0%)
Query 16 PSSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQR 75
P K++LATNIAE+SIT + V+D VK K + T + L + S+A+ QR
Sbjct 314 PPKGTRKVVLATNIAETSITIPGIVYVIDCGYVKVKWYNPKTCSDSLVIVPVSKASAIQR 373
Query 76 RGRTGRVRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMA 121
GR GR+R G L + Y +L P PE++R L ++ + A
Sbjct 374 AGRAGRMRPGKVYRLYTKSDYEALAPRQPPEMRRSELSGAILQLKA 419
> 7303231
Length=1242
Score = 61.2 bits (147), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 36/92 (39%), Positives = 52/92 (56%), Gaps = 1/92 (1%)
Query 22 KIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGR 81
K+++ATNIAE+S+T + V+D VK+K ++ T L + S+AA QR GR GR
Sbjct 855 KVVIATNIAETSLTIDGIFYVVDPGFVKQKVYNSKTGMDSLVVTPISQAAAKQRAGRAGR 914
Query 82 VRDG-TYLCLIPRELYNSLEPFPLPELQRLPL 112
G TY R + + P P+PE+QR L
Sbjct 915 TGPGKTYRLYTERAYRDEMLPTPVPEIQRTNL 946
> Hs20544129
Length=703
Score = 61.2 bits (147), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 37/105 (35%), Positives = 57/105 (54%), Gaps = 0/105 (0%)
Query 17 SSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRR 76
S + K+I+ATN+AE+SIT + V+D VK + + T L + S+A+ +QR
Sbjct 332 SRSVRKVIVATNVAETSITISGIVYVIDCGFVKLRAYNPRTAIECLVVVPVSQASANQRA 391
Query 77 GRTGRVRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMA 121
GR GR R G L E ++ L +PE+QR L V++ + A
Sbjct 392 GRGGRSRSGKCYRLYTEEAFDKLPQSTVPEMQRSNLAPVILQLKA 436
> CE15592
Length=1008
Score = 60.8 bits (146), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 36/97 (37%), Positives = 53/97 (54%), Gaps = 1/97 (1%)
Query 22 KIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGR 81
K++LATNIAE+S+T + V+D K+ A + L + S+AA +QR GR GR
Sbjct 635 KVVLATNIAETSVTIDGINYVIDPGFSKQNSFDARSGVEHLHVVTISKAAANQRAGRAGR 694
Query 82 VRDGTYLCLIPRELY-NSLEPFPLPELQRLPLEDVLV 117
G L Y + LE P+PE+QR L +V++
Sbjct 695 TGPGKCFRLYTAWAYKHELEEQPIPEIQRTNLGNVVL 731
> At1g26370
Length=710
Score = 60.8 bits (146), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 40/114 (35%), Positives = 58/114 (50%), Gaps = 1/114 (0%)
Query 9 KRAKQHAPSSTCM-KIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWA 67
++ K AP+ T K+ILATNIAE+SIT + V+D VK + L + A
Sbjct 331 QQMKVFAPAPTGFRKVILATNIAETSITIPGIRYVIDPGFVKARSYDPSKGMESLDVVPA 390
Query 68 SRAALDQRRGRTGRVRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMA 121
S+A QR GR GR G L P + LE PE++R L ++++ + A
Sbjct 391 SKAQTLQRSGRAGREGPGKSFRLYPEREFEKLEDSTKPEIKRCNLSNIILQLKA 444
> CE01889
Length=1200
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 36/101 (35%), Positives = 54/101 (53%), Gaps = 1/101 (0%)
Query 22 KIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGR 81
K+++ATNIAE+S+T + V+D VK+K + + L + S+AA QR GR GR
Sbjct 809 KVVIATNIAETSLTIDGIFYVVDPGFVKQKIYNPKSGMDSLVVTPISQAAAKQRSGRAGR 868
Query 82 VRDGT-YLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMA 121
G Y R + + P P+PE+QR L L+ + A
Sbjct 869 TGPGKCYRLYTERAFRDEMLPTPVPEIQRTNLASTLLQLKA 909
> CE13245
Length=1019
Score = 60.1 bits (144), Expect = 1e-09, Method: Composition-based stats.
Identities = 35/103 (33%), Positives = 54/103 (52%), Gaps = 0/103 (0%)
Query 18 STCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRG 77
S K+I++TNIAE+S+T + +V+D VK K A + L++ S+A QR G
Sbjct 600 SGARKVIISTNIAETSVTIPGIRVVIDSGKVKTKRFEAFNRIDVLKVHNVSKAQAKQRAG 659
Query 78 RTGRVRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIM 120
R GR G L RE ++ E +PE+ R L +++M
Sbjct 660 RAGRDAPGKCYRLYSREDFHKFEAENMPEILRCNLSATFLELM 702
> Hs20336302
Length=707
Score = 58.9 bits (141), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 36/112 (32%), Positives = 57/112 (50%), Gaps = 1/112 (0%)
Query 10 RAKQHAPSSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASR 69
R Q AP K+I++TNIAE+SIT + V+D +VK K + + L ++ S+
Sbjct 338 RVFQGAPKGY-RKVIISTNIAETSITITGIKYVVDTGMVKAKKYNPDSGLEVLAVQRVSK 396
Query 70 AALDQRRGRTGRVRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMA 121
QR GR GR G L + + + +PE+QR L V++ ++A
Sbjct 397 TQAWQRTGRAGREDSGICYRLYTEDEFEKFDKMTVPEIQRCNLASVMLQLLA 448
> Hs20555648
Length=1041
Score = 58.9 bits (141), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 36/97 (37%), Positives = 54/97 (55%), Gaps = 1/97 (1%)
Query 22 KIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGR 81
K+++ATNIAE+S+T + VLD K+K + T L + S+A+ +QR GR GR
Sbjct 669 KVVVATNIAETSLTIEGIIYVLDPGFCKQKSYNPRTGMESLTVTPCSKASANQRAGRAGR 728
Query 82 VRDGTYLCLIPRELY-NSLEPFPLPELQRLPLEDVLV 117
V G L Y + LE +PE+QR L +V++
Sbjct 729 VAAGKCFRLYTAWAYQHELEETTVPEIQRTSLGNVVL 765
> CE27603
Length=974
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 30/100 (30%), Positives = 55/100 (55%), Gaps = 0/100 (0%)
Query 22 KIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGR 81
K IL+TN+AE+S+T + V+D V T +L W S+A+ +QR+GR GR
Sbjct 367 KCILSTNVAETSVTIDGIRFVIDSGKVNLIKHEPGTGTQKLTEFWVSKASANQRKGRAGR 426
Query 82 VRDGTYLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMA 121
G L +E + ++ F + E+ R+ L+++ + +++
Sbjct 427 TGPGICYRLYSQEQFEKMDDFTVSEINRVSLQEMALKMIS 466
> 7290296
Length=1192
Score = 58.5 bits (140), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 31/101 (30%), Positives = 55/101 (54%), Gaps = 1/101 (0%)
Query 20 CMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRT 79
C +++TN+AE+S+T + V+D K + +T + + S+A+ DQR GR
Sbjct 642 CRLCVVSTNVAETSLTIPHIKYVVDCGRQKTRLYDKLTGVSAFVVTYTSKASADQRAGRA 701
Query 80 GRVRDGTYLCLIPRELYNS-LEPFPLPELQRLPLEDVLVDI 119
GR+ G L +YN E F P++Q+ P+ED+++ +
Sbjct 702 GRISAGHCYRLYSSAVYNDCFEDFSQPDIQKKPVEDLMLQM 742
> YMR128w
Length=1267
Score = 58.5 bits (140), Expect = 4e-09, Method: Composition-based stats.
Identities = 35/116 (30%), Positives = 60/116 (51%), Gaps = 2/116 (1%)
Query 5 TGDLKRAKQHAPSSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQL 64
T + R Q P + + I+ATN+AE+S+T V V+D KE+ + ++
Sbjct 740 TKEQMRVFQKPPQGSRL-CIVATNVAETSLTIPGVRYVVDSGRSKERKYNESNGVQSFEV 798
Query 65 RWASRAALDQRRGRTGRVRDGTYLCLIPRELY-NSLEPFPLPELQRLPLEDVLVDI 119
W S+A+ +QR GR GR G L ++ + E F PE+ R+P+E +++ +
Sbjct 799 GWVSKASANQRSGRAGRTGPGHCYRLYSSAVFEHDFEQFSKPEILRMPVESIVLQM 854
> At1g33390
Length=1237
Score = 58.5 bits (140), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 31/96 (32%), Positives = 54/96 (56%), Gaps = 1/96 (1%)
Query 23 IILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGRV 82
+++ATN+AE+S+T + V+D VK K+ + T ++ W S+A+ QR GR GR
Sbjct 675 VVVATNVAETSLTIPGIKYVVDTGRVKVKNYDSKTGMESYEVDWISQASASQRAGRAGRT 734
Query 83 RDGTYLCLIPRELY-NSLEPFPLPELQRLPLEDVLV 117
G L ++ N E LPE+ ++P++ V++
Sbjct 735 GPGHCYRLYSSAVFSNIFEESSLPEIMKVPVDGVIL 770
> 7298547
Length=894
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 35/101 (34%), Positives = 57/101 (56%), Gaps = 1/101 (0%)
Query 22 KIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGR 81
K+ILATNIAE+S+T ++ V+D K+ + ++ T L + S+A+ +QR GR GR
Sbjct 522 KVILATNIAETSLTIDNIIYVIDPGFAKQNNFNSRTGMESLMVVPISKASANQRAGRAGR 581
Query 82 VRDGTYLCLIPRELY-NSLEPFPLPELQRLPLEDVLVDIMA 121
G L Y + LE +PE+QR+ L + ++ + A
Sbjct 582 TAPGKCFRLYTAWAYKHELEDNTVPEIQRINLGNAVLMLKA 622
> ECU09g0150
Length=784
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 35/97 (36%), Positives = 54/97 (55%), Gaps = 2/97 (2%)
Query 22 KIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGR 81
K I++TN+AE+S+T ++ V+D L K V++ L SRA DQR GR GR
Sbjct 374 KCIVSTNVAETSLTIPNIGYVIDTGLQK-ISVYSYDTGESLVTVPISRANADQRTGRAGR 432
Query 82 VRDGTYLCLIPRELY-NSLEPFPLPELQRLPLEDVLV 117
R G + + Y N + P P+PE+QR + +V++
Sbjct 433 TRPGVCYRMYTADTYENDMLPSPVPEIQRTNIHNVVL 469
> At2g35340
Length=1087
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 36/101 (35%), Positives = 52/101 (51%), Gaps = 1/101 (0%)
Query 22 KIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGR 81
K++LATNIAE+S+T + V+D K K + T L + S+A+ QR GR GR
Sbjct 716 KVVLATNIAETSLTIDGIKYVVDPGFSKMKSYNPRTGMESLLVTPISKASATQRTGRAGR 775
Query 82 VRDGT-YLCLIPRELYNSLEPFPLPELQRLPLEDVLVDIMA 121
G Y YN LE +PE+QR L V++ + +
Sbjct 776 TSPGKCYRLYTAFNYYNDLEDNTVPEIQRTNLASVVLSLKS 816
> Hs4503293
Length=1041
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 36/97 (37%), Positives = 53/97 (54%), Gaps = 1/97 (1%)
Query 22 KIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGR 81
K+++ATNIAE+S T + VLD K+K + T L + S+A+ +QR GR GR
Sbjct 669 KVVVATNIAETSPTIEGIIYVLDPGFCKQKSYNPRTGMESLTVTPCSKASANQRAGRAGR 728
Query 82 VRDGTYLCLIPRELY-NSLEPFPLPELQRLPLEDVLV 117
V G L Y + LE +PE+QR L +V++
Sbjct 729 VAAGKCFRLYTAWAYQHELEETTVPEIQRTSLGNVVL 765
> At1g27900
Length=708
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 33/98 (33%), Positives = 50/98 (51%), Gaps = 1/98 (1%)
Query 16 PSSTCMKIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQR 75
P C + I++TNIAE+S+T V V+D VK++ + + L + S+ +QR
Sbjct 269 PPPNCRRFIVSTNIAETSLTVDGVVYVIDSGYVKQRQYNPSSGMFSLDVIQISKVQANQR 328
Query 76 RGRTGRVRDGTYLCLIPRELY-NSLEPFPLPELQRLPL 112
GR GR R G L P +Y + +PE+QR L
Sbjct 329 AGRAGRTRPGKCYRLYPLAVYRDDFLDATIPEIQRTSL 366
> ECU11g0860
Length=664
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 32/86 (37%), Positives = 45/86 (52%), Gaps = 0/86 (0%)
Query 22 KIILATNIAESSITFVDVCIVLDFALVKEKHVHAVTKAHRLQLRWASRAALDQRRGRTGR 81
KI+LATNIAE+SIT V V+D K+ A L++ W S+A QR GR GR
Sbjct 314 KIVLATNIAETSITIEGVRYVVDSGRAKQMRYSASFGMDILEVVWISKAQAKQRAGRAGR 373
Query 82 VRDGTYLCLIPRELYNSLEPFPLPEL 107
+ G + +E Y ++ PE+
Sbjct 374 TQAGKVFRMYSKEEYQKMDDNTTPEI 399
Lambda K H
0.322 0.136 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1821716300
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40