bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
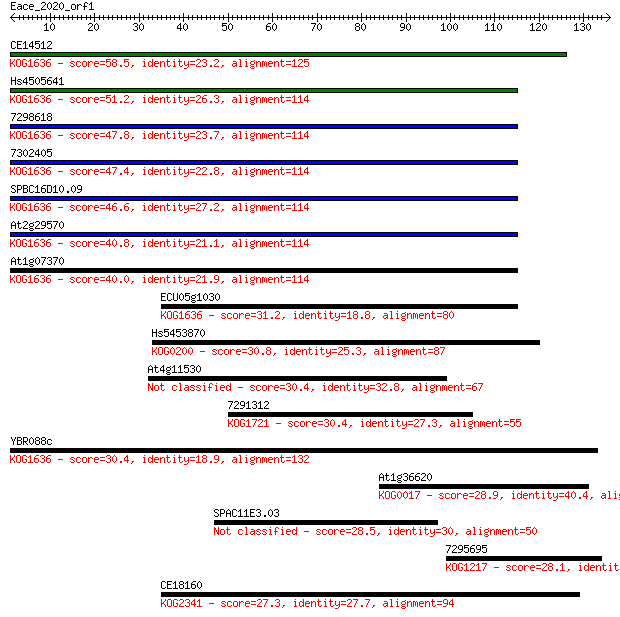
Query= Eace_2020_orf1
Length=135
Score E
Sequences producing significant alignments: (Bits) Value
CE14512 58.5 3e-09
Hs4505641 51.2 6e-07
7298618 47.8 7e-06
7302405 47.4 7e-06
SPBC16D10.09 46.6 1e-05
At2g29570 40.8 8e-04
At1g07370 40.0 0.001
ECU05g1030 31.2 0.60
Hs5453870 30.8 0.81
At4g11530 30.4 0.98
7291312 30.4 0.99
YBR088c 30.4 1.1
At1g36620 28.9 2.9
SPAC11E3.03 28.5 3.7
7295695 28.1 5.3
CE18160 27.3 9.3
> CE14512
Length=263
Score = 58.5 bits (140), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 29/125 (23%), Positives = 67/125 (53%), Gaps = 5/125 (4%)
Query 1 VTLGLSVATILKAIQPVESNKHIVHLSTLKGSDEDEEVLHIAIEDPERKDNWHMEISLLT 60
+ LGLS+A + KA++ ++ + LK + + + + DP+R + + ++
Sbjct 66 INLGLSLANMSKALKCANNDDTCM----LKYEENEGDSIIFTFADPKRDKTQDVTVKMMD 121
Query 61 VAEERLQVPNEPYACLCTVDAKEFQELLRYIHSIAESVRIRCEKDSLILSAKGENLAVTR 120
+ E L +P++ YA +C + A EFQ+ + + + ++S+ I K ++ + KG+ + +
Sbjct 122 IDSEHLGIPDQDYAVVCEMPAGEFQKTCKDLSTFSDSLNITATKAGIVFTGKGD-IGSSV 180
Query 121 VIHAP 125
V ++P
Sbjct 181 VTYSP 185
> Hs4505641
Length=261
Score = 51.2 bits (121), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 30/114 (26%), Positives = 62/114 (54%), Gaps = 5/114 (4%)
Query 1 VTLGLSVATILKAIQPVESNKHIVHLSTLKGSDEDEEVLHIAIEDPERKDNWHMEISLLT 60
+ +G+++ ++ K ++ N+ I+ TL+ D + + L + E P ++ E+ L+
Sbjct 66 LAMGVNLTSMSKILK-CAGNEDII---TLRAED-NADTLALVFEAPNQEKVSDYEMKLMD 120
Query 61 VAEERLQVPNEPYACLCTVDAKEFQELLRYIHSIAESVRIRCEKDSLILSAKGE 114
+ E+L +P + Y+C+ + + EF + R + I ++V I C KD + SA GE
Sbjct 121 LDVEQLGIPEQEYSCVVKMPSGEFARICRDLSHIGDAVVISCAKDGVKFSASGE 174
> 7298618
Length=255
Score = 47.8 bits (112), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 27/114 (23%), Positives = 57/114 (50%), Gaps = 5/114 (4%)
Query 1 VTLGLSVATILKAIQPVESNKHIVHLSTLKGSDEDEEVLHIAIEDPERKDNWHMEISLLT 60
V+LGL + ++ K ++ S+ + T+K D E++ ++ E ++ E+ LL
Sbjct 66 VSLGLDLKSLGKVLKCANSDDAV----TIKAVDRPEKI-TLSFESDGKERTADYELKLLN 120
Query 61 VAEERLQVPNEPYACLCTVDAKEFQELLRYIHSIAESVRIRCEKDSLILSAKGE 114
+ ++ +++P + Y C + + EF + R + ES+ I C + AKG+
Sbjct 121 LDQDHMEIPKKDYTCFIQLPSSEFARICRDMSMFDESLTIACSSKGIRFLAKGD 174
> 7302405
Length=260
Score = 47.4 bits (111), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 26/114 (22%), Positives = 60/114 (52%), Gaps = 5/114 (4%)
Query 1 VTLGLSVATILKAIQPVESNKHIVHLSTLKGSDEDEEVLHIAIEDPERKDNWHMEISLLT 60
+++G+++ ++ K ++ + ++ T+K D + + + I E ++ E+ L+
Sbjct 66 LSMGMNLGSMAKILKCANNEDNV----TMKAQD-NADTVTIMFESANQEKVSDYEMKLMN 120
Query 61 VAEERLQVPNEPYACLCTVDAKEFQELLRYIHSIAESVRIRCEKDSLILSAKGE 114
+ +E L +P ++C+ + A EF + R + +ESV I C K+ + SA G+
Sbjct 121 LDQEHLGIPETDFSCVVRMPAMEFARICRDLAQFSESVVICCTKEGVKFSASGD 174
> SPBC16D10.09
Length=260
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 31/114 (27%), Positives = 61/114 (53%), Gaps = 5/114 (4%)
Query 1 VTLGLSVATILKAIQPVESNKHIVHLSTLKGSDEDEEVLHIAIEDPERKDNWHMEISLLT 60
+ LG+++ + K ++ + N+ +V TLK D EVL++ E + ++ L+
Sbjct 66 IALGINLNALSKVLRCAQ-NEDLV---TLKAEDT-PEVLNLVFESEKNDRISDYDVKLMD 120
Query 61 VAEERLQVPNEPYACLCTVDAKEFQELLRYIHSIAESVRIRCEKDSLILSAKGE 114
+ +E L +P+ Y T+ A EFQ + R + ++++SV I K+ + S KG+
Sbjct 121 IDQEHLGIPDIEYDATITMPAAEFQRITRDLLTLSDSVTINASKEGVRFSCKGD 174
> At2g29570
Length=264
Score = 40.8 bits (94), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 24/114 (21%), Positives = 53/114 (46%), Gaps = 5/114 (4%)
Query 1 VTLGLSVATILKAIQPVESNKHIVHLSTLKGSDEDEEVLHIAIEDPERKDNWHMEISLLT 60
+++G+++ + K ++ N I+ + GSD + E P + E+ L+
Sbjct 66 LSMGMNLGNMSKMLK-CAGNDDIITIKADDGSD----TVTFMFESPTQDKIADFEMKLMD 120
Query 61 VAEERLQVPNEPYACLCTVDAKEFQELLRYIHSIAESVRIRCEKDSLILSAKGE 114
+ E L +P+ Y + + + EF + + + SI ++V I K+ + S G+
Sbjct 121 IDSEHLGIPDAEYHSIVRMPSGEFSRICKDLSSIGDTVVISVTKEGVKFSTAGD 174
> At1g07370
Length=263
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/114 (21%), Positives = 55/114 (48%), Gaps = 5/114 (4%)
Query 1 VTLGLSVATILKAIQPVESNKHIVHLSTLKGSDEDEEVLHIAIEDPERKDNWHMEISLLT 60
+++G+++ + K ++ N I+ T+K D + V + E P + E+ L+
Sbjct 66 LSMGMNLGNMSKMLK-CAGNDDII---TIKADDGGDTVTFM-FESPTQDKIADFEMKLMD 120
Query 61 VAEERLQVPNEPYACLCTVDAKEFQELLRYIHSIAESVRIRCEKDSLILSAKGE 114
+ E L +P+ Y + + + EF + + + SI ++V I K+ + S G+
Sbjct 121 IDSEHLGIPDAEYHSIVRMPSNEFSRICKDLSSIGDTVVISVTKEGVKFSTAGD 174
> ECU05g1030
Length=267
Score = 31.2 bits (69), Expect = 0.60, Method: Compositional matrix adjust.
Identities = 15/80 (18%), Positives = 34/80 (42%), Gaps = 0/80 (0%)
Query 35 DEEVLHIAIEDPERKDNWHMEISLLTVAEERLQVPNEPYACLCTVDAKEFQELLRYIHSI 94
D+ ++ I + + ++ L T E +P + T+ A EF + + + +
Sbjct 107 DDATTNLNIRNTREGNVLSFKLKLFTSDSEAYNIPEFDFDASATIPADEFMYVPKLVGTF 166
Query 95 AESVRIRCEKDSLILSAKGE 114
+ V ++ E +L+ GE
Sbjct 167 GDVVGVQAEGKTLMFFQTGE 186
> Hs5453870
Length=1089
Score = 30.8 bits (68), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 22/93 (23%), Positives = 41/93 (44%), Gaps = 6/93 (6%)
Query 33 DEDEEVLHIAIEDPERKDNWHMEISLLTVAEERLQVPNE-----PYACLCTVDAKEFQEL 87
D+D ++ DPE H ++ + + Q N PY C TV K+FQ +
Sbjct 142 DDDSAIIPCRTTDPETPVTLHNSEGVVPASYDSRQGFNGTFTVGPYICEATVKGKKFQTI 201
Query 88 LRYIHSIAESVRIRCEKDSL-ILSAKGENLAVT 119
++++ + + E ++L + GE + VT
Sbjct 202 PFNVYALKATSELDLEMEALKTVYKSGETIVVT 234
> At4g11530
Length=931
Score = 30.4 bits (67), Expect = 0.98, Method: Composition-based stats.
Identities = 22/71 (30%), Positives = 32/71 (45%), Gaps = 6/71 (8%)
Query 32 SDEDEEVLHIAI----EDPERKDNWHMEISLLTVAEERLQVPNEPYACLCTVDAKEFQEL 87
S E +HIA+ EDP + I +LT + L VP P CL D + Q+
Sbjct 848 SSEATRCIHIALLCVQEDPADRPLLPAIIMMLTSSTTTLHVPRAPGFCLSGRDLE--QDG 905
Query 88 LRYIHSIAESV 98
+ Y S + S+
Sbjct 906 VEYTESTSRSI 916
> 7291312
Length=835
Score = 30.4 bits (67), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 15/58 (25%), Positives = 33/58 (56%), Gaps = 3/58 (5%)
Query 50 DNWHMEISLLT--VAEERLQVPNEPYAC-LCTVDAKEFQELLRYIHSIAESVRIRCEK 104
DN+ ++ +L+T + + + + Y C LC+ + ++F+ L ++HS +R C+K
Sbjct 600 DNYEIDANLVTEFIRQHTSPLGSGRYICHLCSTEFRQFKGLQNHMHSHTNWIRANCKK 657
> YBR088c
Length=258
Score = 30.4 bits (67), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 25/134 (18%), Positives = 64/134 (47%), Gaps = 8/134 (5%)
Query 1 VTLGLSVATILKAIQPVESNKHIVHLSTLKGSDEDEEVLHIAIEDPERKDNWHMEISLLT 60
VTLG+ + ++ K ++ + + ++ D + + + ED ++ + L+
Sbjct 66 VTLGMDLTSLSKILRCGNNTDTLTLIA-----DNTPDSIILLFEDTKKDRIAEYSLKLMD 120
Query 61 VAEERLQVPNEPYACLCTVDAKEFQELLRYIHSIAESVRIRCEKDSLILSAKGENLAVTR 120
+ + L++ Y ++ + EF +++R + +++S+ I K+++ A G ++
Sbjct 121 IDADFLKIEELQYDSTLSLPSSEFSKIVRDLSQLSDSINIMITKETIKFVADG-DIGSGS 179
Query 121 VIHAP--DMQCSES 132
VI P DM+ E+
Sbjct 180 VIIKPFVDMEHPET 193
> At1g36620
Length=1152
Score = 28.9 bits (63), Expect = 2.9, Method: Composition-based stats.
Identities = 19/55 (34%), Positives = 28/55 (50%), Gaps = 11/55 (20%)
Query 84 FQELLRYIHSIAESV--------RIRCEKDSLILSAKGENLAVTRVIHAPDMQCS 130
F +LL I+ I SV I +K +LIL G N+ +T V+ PD+ C+
Sbjct 396 FLDLLTEIYEIPHSVVVLPNAKHTIATKKGTLIL---GANMKLTHVLFVPDLSCT 447
> SPAC11E3.03
Length=257
Score = 28.5 bits (62), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 15/50 (30%), Positives = 26/50 (52%), Gaps = 0/50 (0%)
Query 47 ERKDNWHMEISLLTVAEERLQVPNEPYACLCTVDAKEFQELLRYIHSIAE 96
E ++ H+ S ++ ++LQ+PN + L +EF EL + I I E
Sbjct 55 ENENELHINNSGMSELNKKLQLPNVELSTLSHTQEQEFNELNKLIRKINE 104
> 7295695
Length=3680
Score = 28.1 bits (61), Expect = 5.3, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 16/35 (45%), Gaps = 0/35 (0%)
Query 99 RIRCEKDSLILSAKGENLAVTRVIHAPDMQCSESF 133
R +CE L +A G N HAPD C E F
Sbjct 2461 RAKCESVCLGRAACGRNAECVARSHAPDCLCKEGF 2495
> CE18160
Length=549
Score = 27.3 bits (59), Expect = 9.3, Method: Composition-based stats.
Identities = 26/94 (27%), Positives = 40/94 (42%), Gaps = 1/94 (1%)
Query 35 DEEVLHIAIEDPERKDNWHMEISLLTVAEERLQVPNEPYACLCTVDAKEFQELLRYIHSI 94
+EE L +AI D + + VAE R++ P + D K L +
Sbjct 375 EEEAL-VAISDAVESHLRELITLMAGVAEHRVESLRIPENYVAIDDVKRQLRFLEDLDRQ 433
Query 95 AESVRIRCEKDSLILSAKGENLAVTRVIHAPDMQ 128
E +R EK+SLI +K +N + A +MQ
Sbjct 434 EEELRESREKESLIRMSKNKNSGKETIEKAKEMQ 467
Lambda K H
0.316 0.131 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1388795348
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40