bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1993_orf1
Length=113
Score E
Sequences producing significant alignments: (Bits) Value
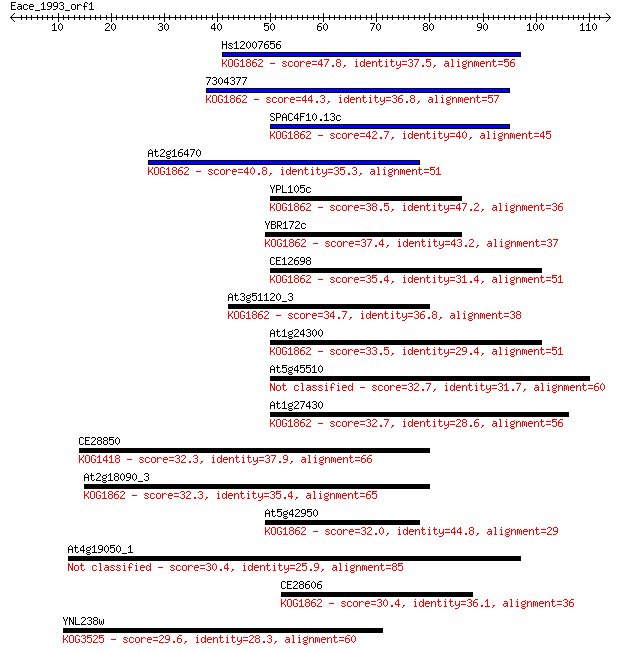
Hs12007656 47.8 5e-06
7304377 44.3 6e-05
SPAC4F10.13c 42.7 1e-04
At2g16470 40.8 6e-04
YPL105c 38.5 0.003
YBR172c 37.4 0.006
CE12698 35.4 0.023
At3g51120_3 34.7 0.045
At1g24300 33.5 0.098
At5g45510 32.7 0.15
At1g27430 32.7 0.19
CE28850 32.3 0.20
At2g18090_3 32.3 0.22
At5g42950 32.0 0.32
At4g19050_1 30.4 0.81
CE28606 30.4 0.86
YNL238w 29.6 1.4
> Hs12007656
Length=817
Score = 47.8 bits (112), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 32/56 (57%), Gaps = 0/56 (0%)
Query 41 PQENEAAHSWCYLDAFNKCQGPFSTLTMLQWLELGYFDPTRPLRRDDEKGFTPLHD 96
P + AA W Y D + QGPF+T M +W + GYF + ++R ++GF PL +
Sbjct 346 PLSHGAARKWFYKDPQGEIQGPFTTQEMAEWFQAGYFSMSLLVKRGCDEGFQPLGE 401
> 7304377
Length=1465
Score = 44.3 bits (103), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 21/57 (36%), Positives = 28/57 (49%), Gaps = 3/57 (5%)
Query 38 LLDPQENEAAHSWCYLDAFNKCQGPFSTLTMLQWLELGYFDPTRPLRRDDEKGFTPL 94
++ P NE W Y D QGPFS + M +W GYF+ +RR + F PL
Sbjct 553 IITPNLNEL---WFYRDPQANVQGPFSAVEMTEWYRAGYFNENLFVRRYSDNRFRPL 606
> SPAC4F10.13c
Length=992
Score = 42.7 bits (99), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 18/45 (40%), Positives = 25/45 (55%), Gaps = 0/45 (0%)
Query 50 WCYLDAFNKCQGPFSTLTMLQWLELGYFDPTRPLRRDDEKGFTPL 94
W Y D N QGPF+ + M QW GYF P++R +E+ + L
Sbjct 389 WLYKDPQNNVQGPFTGVDMHQWYRAGYFPLGLPIKRLEEEEYYSL 433
> At2g16470
Length=670
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 27/54 (50%), Gaps = 3/54 (5%)
Query 27 LLLELSRVPERLLD---PQENEAAHSWCYLDAFNKCQGPFSTLTMLQWLELGYF 77
+ E+S P ++ P+ N++ W Y D K QGPFS + +W GYF
Sbjct 170 IASEISMAPPAVVSQPVPKSNDSEKIWHYKDPSGKVQGPFSMAQLRKWNNTGYF 223
> YPL105c
Length=849
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 17/36 (47%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 50 WCYLDAFNKCQGPFSTLTMLQWLELGYFDPTRPLRR 85
W Y+D+ QGPF T M QW + GYF PT + R
Sbjct 152 WKYIDSNGNIQGPFGTNNMSQWYQGGYFTPTLQICR 187
> YBR172c
Length=790
Score = 37.4 bits (85), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 49 SWCYLDAFNKCQGPFSTLTMLQWLELGYFDPTRPLRR 85
SW Y+D + GPF+T M QW GYF T + R
Sbjct 257 SWRYIDTQGQIHGPFTTQMMSQWYIGGYFASTLQISR 293
> CE12698
Length=245
Score = 35.4 bits (80), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 29/51 (56%), Gaps = 0/51 (0%)
Query 50 WCYLDAFNKCQGPFSTLTMLQWLELGYFDPTRPLRRDDEKGFTPLHDGSRI 100
W YL ++ GP+ + ML WL+ GYF+ L+ ++E + L + S++
Sbjct 15 WHYLGPDSEKYGPYMSKDMLFWLQAGYFNDGLQLKTENEPNYHTLGEWSQL 65
> At3g51120_3
Length=693
Score = 34.7 bits (78), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 42 QENEAAHSWCYLDAFNKCQGPFSTLTMLQWLELGYFDP 79
+++E + W Y D K QGPFS + + +W G+F P
Sbjct 112 KDDEESEIWHYRDPTGKTQGPFSMVQLRRWKFSGHFPP 149
> At1g24300
Length=1417
Score = 33.5 bits (75), Expect = 0.098, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 23/51 (45%), Gaps = 0/51 (0%)
Query 50 WCYLDAFNKCQGPFSTLTMLQWLELGYFDPTRPLRRDDEKGFTPLHDGSRI 100
+ Y+D QGPF ++ W E G+F +R + TP D R+
Sbjct 401 FLYIDPQGVIQGPFIGSDIISWFEQGFFGTDLQVRLANAPEGTPFQDLGRV 451
> At5g45510
Length=1214
Score = 32.7 bits (73), Expect = 0.15, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 29/63 (46%), Gaps = 3/63 (4%)
Query 50 WCYLDAFNKCQGPFSTLTMLQWLELGYFDPTRPLRRDDEKG---FTPLHDGSRIKRAAKD 106
W LD F C + + QW+ GYFDP R + + + G F L D +K +
Sbjct 467 WHSLDFFEHCGCVYYRDLITQWILEGYFDPVRSVEKAYQDGHSIFMELIDRGMLKIQENN 526
Query 107 IVL 109
+V+
Sbjct 527 VVV 529
> At1g27430
Length=1453
Score = 32.7 bits (73), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 16/56 (28%), Positives = 23/56 (41%), Gaps = 0/56 (0%)
Query 50 WCYLDAFNKCQGPFSTLTMLQWLELGYFDPTRPLRRDDEKGFTPLHDGSRIKRAAK 105
+ Y+D QGPF ++ W E G+F +R TP D R+ K
Sbjct 401 FLYIDPQGVIQGPFIGSDIISWFEQGFFGTDLQVRLASAPEGTPFQDLGRVMSYIK 456
> CE28850
Length=411
Score = 32.3 bits (72), Expect = 0.20, Method: Composition-based stats.
Identities = 25/66 (37%), Positives = 33/66 (50%), Gaps = 10/66 (15%)
Query 14 HSQQHGVGRVKSSLLLELSRVPERLLDPQENEAAHSWCYLDAFNKCQGPFSTLTMLQWLE 73
H Q VG V +SLLL + +P + E + W YLDAF C F +LT +
Sbjct 258 HIQLIHVGVVFASLLLFVFAIPAWVFSSIETD----WSYLDAFYYC---FVSLTTIG--- 307
Query 74 LGYFDP 79
LG F+P
Sbjct 308 LGDFEP 313
> At2g18090_3
Length=394
Score = 32.3 bits (72), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 26/65 (40%), Gaps = 7/65 (10%)
Query 15 SQQHGVGRVKSSLLLELSRVPERLLDPQENEAAHSWCYLDAFNKCQGPFSTLTMLQWLEL 74
S+ H V VK LS P LD W Y D K GPFS + QW
Sbjct 1 SRNHAV--VKPDTSATLSNKPIDGLDTN-----MVWLYGDPDGKIHGPFSLYNLRQWNSS 53
Query 75 GYFDP 79
G+F P
Sbjct 54 GHFPP 58
> At5g42950
Length=1714
Score = 32.0 bits (71), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 15/29 (51%), Gaps = 0/29 (0%)
Query 49 SWCYLDAFNKCQGPFSTLTMLQWLELGYF 77
S Y D QGPFS ++ W E GYF
Sbjct 548 SLYYKDPQGLIQGPFSGSDIIGWFEAGYF 576
> At4g19050_1
Length=1171
Score = 30.4 bits (67), Expect = 0.81, Method: Composition-based stats.
Identities = 22/89 (24%), Positives = 41/89 (46%), Gaps = 7/89 (7%)
Query 12 EKHSQQHGVGRVKSSLLLELSRVPERLLDPQENEA----AHSWCYLDAFNKCQGPFSTLT 67
E + ++ + R + + +L+LS LL P E A W LD + +
Sbjct 263 EAAASKNAIDRSRYNPVLQLSY---ELLKPDETVKRPVIACFWHILDFYKYSGCAYYRDL 319
Query 68 MLQWLELGYFDPTRPLRRDDEKGFTPLHD 96
++ W+ GYFDP + + + ++G + L D
Sbjct 320 IVHWMLEGYFDPVKSVDKAYQEGHSILMD 348
> CE28606
Length=717
Score = 30.4 bits (67), Expect = 0.86, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 17/36 (47%), Gaps = 0/36 (0%)
Query 52 YLDAFNKCQGPFSTLTMLQWLELGYFDPTRPLRRDD 87
Y D QGP+ T+L W + GYF +R D
Sbjct 400 YTDDRGTVQGPYGASTVLDWYQKGYFSDNHQMRFTD 435
> YNL238w
Length=814
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 29/60 (48%), Gaps = 7/60 (11%)
Query 11 AEKHSQQHGVGRVKSSLLLELSRVPERLLDPQENEAAHSWCYLDAFNKCQGPFSTLTMLQ 70
+K+S ++G G++ + L+E+S+ EN A +W YL Q ST L+
Sbjct 435 GKKYSHRYGFGKIDAHKLIEMSKT-------WENVNAQTWFYLPTLYVSQSTNSTEETLE 487
Lambda K H
0.317 0.132 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1185472426
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40