bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1917_orf1
Length=91
Score E
Sequences producing significant alignments: (Bits) Value
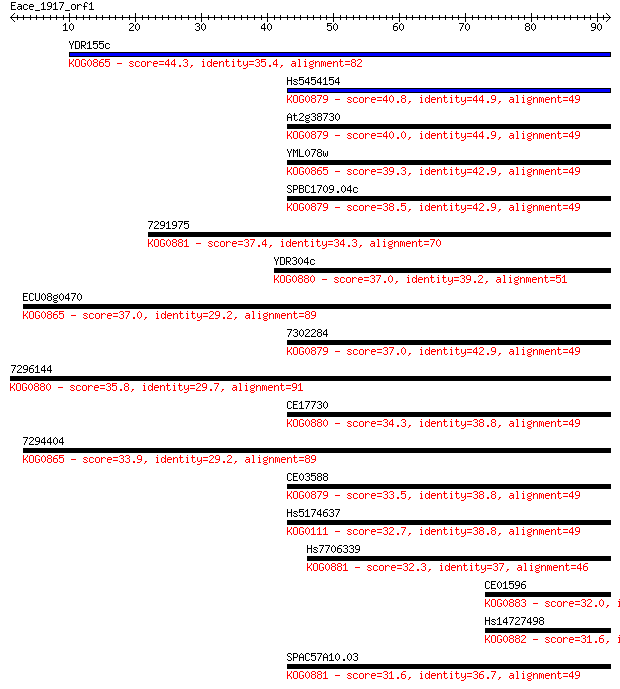
YDR155c 44.3 5e-05
Hs5454154 40.8 6e-04
At2g38730 40.0 0.001
YML078w 39.3 0.002
SPBC1709.04c 38.5 0.003
7291975 37.4 0.007
YDR304c 37.0 0.008
ECU08g0470 37.0 0.010
7302284 37.0 0.010
7296144 35.8 0.018
CE17730 34.3 0.060
7294404 33.9 0.066
CE03588 33.5 0.091
Hs5174637 32.7 0.17
Hs7706339 32.3 0.25
CE01596 32.0 0.30
Hs14727498 31.6 0.37
SPAC57A10.03 31.6 0.40
> YDR155c
Length=162
Score = 44.3 bits (103), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 32/89 (35%), Gaps = 24/89 (26%)
Query 10 PLESAAMPETIQGLSGAAATAAAAKAIAAGCVP-------HFRAGLLCMANSAAASSKQQ 62
P +QG A K+I G P H R GLL MAN+
Sbjct 50 PFHRVIPDFMLQGGDFTAGNGTGGKSIYGGKFPDENFKKHHDRPGLLSMANAGPN----- 104
Query 63 QQQQQEQQQRSGTNGCQFFITCGPCLWLD 91
TNG QFFIT PC WLD
Sbjct 105 ------------TNGSQFFITTVPCPWLD 121
> Hs5454154
Length=177
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 22/49 (44%), Positives = 23/49 (46%), Gaps = 17/49 (34%)
Query 43 HFRAGLLCMANSAAASSKQQQQQQQEQQQRSGTNGCQFFITCGPCLWLD 91
H GLL MANS + TNGCQFFITC C WLD
Sbjct 104 HSAPGLLSMANSGPS-----------------TNGCQFFITCSKCDWLD 135
> At2g38730
Length=199
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/49 (44%), Positives = 22/49 (44%), Gaps = 17/49 (34%)
Query 43 HFRAGLLCMANSAAASSKQQQQQQQEQQQRSGTNGCQFFITCGPCLWLD 91
H GLL MANS TNGCQFFITC C WLD
Sbjct 125 HTGPGLLSMANSGP-----------------NTNGCQFFITCAKCDWLD 156
> YML078w
Length=182
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 21/49 (42%), Positives = 23/49 (46%), Gaps = 17/49 (34%)
Query 43 HFRAGLLCMANSAAASSKQQQQQQQEQQQRSGTNGCQFFITCGPCLWLD 91
H +AGLL MAN+ TNG QFFIT PC WLD
Sbjct 110 HDKAGLLSMANAGP-----------------NTNGSQFFITTVPCPWLD 141
> SPBC1709.04c
Length=173
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 21/49 (42%), Positives = 23/49 (46%), Gaps = 17/49 (34%)
Query 43 HFRAGLLCMANSAAASSKQQQQQQQEQQQRSGTNGCQFFITCGPCLWLD 91
H R GLL MAN+ S NGCQFFIT PC +LD
Sbjct 100 HDRPGLLSMANAGKDS-----------------NGCQFFITTVPCDFLD 131
> 7291975
Length=176
Score = 37.4 bits (85), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 28/70 (40%), Gaps = 17/70 (24%)
Query 22 GLSGAAATAAAAKAIAAGCVPHFRAGLLCMANSAAASSKQQQQQQQEQQQRSGTNGCQFF 81
G GA+ + G + H AG+L MANS TNG QFF
Sbjct 80 GRGGASIYGSEFADELHGDLRHTGAGILSMANSGPD-----------------TNGSQFF 122
Query 82 ITCGPCLWLD 91
IT P WLD
Sbjct 123 ITLAPTQWLD 132
> YDR304c
Length=225
Score = 37.0 bits (84), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 21/51 (41%), Gaps = 17/51 (33%)
Query 41 VPHFRAGLLCMANSAAASSKQQQQQQQEQQQRSGTNGCQFFITCGPCLWLD 91
V H + G L MAN TNG QFFIT PC WLD
Sbjct 121 VKHDKPGRLSMANRG-----------------KNTNGSQFFITTVPCPWLD 154
> ECU08g0470
Length=172
Score = 37.0 bits (84), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 30/89 (33%), Gaps = 42/89 (47%)
Query 3 SIYGSSFPLESAAMPETIQGLSGAAATAAAAKAIAAGCVPHFRAGLLCMANSAAASSKQQ 62
SIYG FP E+ + H + G+L MAN A
Sbjct 80 SIYGEKFPDENFELK-------------------------HTKEGILSMANCGAH----- 109
Query 63 QQQQQEQQQRSGTNGCQFFITCGPCLWLD 91
TNG QFFIT G WLD
Sbjct 110 ------------TNGSQFFITLGKTQWLD 126
> 7302284
Length=183
Score = 37.0 bits (84), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 21/49 (42%), Positives = 22/49 (44%), Gaps = 17/49 (34%)
Query 43 HFRAGLLCMANSAAASSKQQQQQQQEQQQRSGTNGCQFFITCGPCLWLD 91
H GLL MANS TNGCQFFITC C +LD
Sbjct 110 HDSPGLLSMANSGKE-----------------TNGCQFFITCAKCNFLD 141
> 7296144
Length=237
Score = 35.8 bits (81), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 27/91 (29%), Positives = 31/91 (34%), Gaps = 40/91 (43%)
Query 1 SVSIYGSSFPLESAAMPETIQGLSGAAATAAAAKAIAAGCVPHFRAGLLCMANSAAASSK 60
S+SIYG FP E A+ V H R G L MAN
Sbjct 99 SISIYGDYFPDEDKAL-----------------------AVEHNRPGYLGMANRGPD--- 132
Query 61 QQQQQQQEQQQRSGTNGCQFFITCGPCLWLD 91
TNGCQF++T WLD
Sbjct 133 --------------TNGCQFYVTTVGAKWLD 149
> CE17730
Length=204
Score = 34.3 bits (77), Expect = 0.060, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 21/49 (42%), Gaps = 17/49 (34%)
Query 43 HFRAGLLCMANSAAASSKQQQQQQQEQQQRSGTNGCQFFITCGPCLWLD 91
H+ AG L MAN+ A TNG QFFIT WLD
Sbjct 117 HYGAGWLSMANAGAD-----------------TNGSQFFITTVKTPWLD 148
> 7294404
Length=164
Score = 33.9 bits (76), Expect = 0.066, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 31/89 (34%), Gaps = 42/89 (47%)
Query 3 SIYGSSFPLESAAMPETIQGLSGAAATAAAAKAIAAGCVPHFRAGLLCMANSAAASSKQQ 62
SIYG+ FP E+ + H AG+L MAN+ A
Sbjct 77 SIYGNKFPDENFELK-------------------------HTGAGVLSMANAGAN----- 106
Query 63 QQQQQEQQQRSGTNGCQFFITCGPCLWLD 91
TNG QFFI G WLD
Sbjct 107 ------------TNGSQFFICTGKTTWLD 123
> CE03588
Length=183
Score = 33.5 bits (75), Expect = 0.091, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 22/49 (44%), Gaps = 17/49 (34%)
Query 43 HFRAGLLCMANSAAASSKQQQQQQQEQQQRSGTNGCQFFITCGPCLWLD 91
H G+L MAN+ S TNGCQFFITC +LD
Sbjct 110 HIGPGMLSMANAG-----------------SDTNGCQFFITCAKTDFLD 141
> Hs5174637
Length=301
Score = 32.7 bits (73), Expect = 0.17, Method: Composition-based stats.
Identities = 19/49 (38%), Positives = 20/49 (40%), Gaps = 17/49 (34%)
Query 43 HFRAGLLCMANSAAASSKQQQQQQQEQQQRSGTNGCQFFITCGPCLWLD 91
H GLL MANS TNG QFF+TC WLD
Sbjct 228 HTGPGLLSMANSGP-----------------NTNGSQFFLTCDKTDWLD 259
> Hs7706339
Length=166
Score = 32.3 bits (72), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 20/46 (43%), Gaps = 17/46 (36%)
Query 46 AGLLCMANSAAASSKQQQQQQQEQQQRSGTNGCQFFITCGPCLWLD 91
AG+L MAN+ TNG QFF+T P WLD
Sbjct 95 AGILAMANAGPD-----------------TNGSQFFVTLAPTQWLD 123
> CE01596
Length=523
Score = 32.0 bits (71), Expect = 0.30, Method: Composition-based stats.
Identities = 13/19 (68%), Positives = 14/19 (73%), Gaps = 0/19 (0%)
Query 73 SGTNGCQFFITCGPCLWLD 91
S TNG QFFIT PC +LD
Sbjct 373 SNTNGSQFFITFRPCKYLD 391
> Hs14727498
Length=565
Score = 31.6 bits (70), Expect = 0.37, Method: Composition-based stats.
Identities = 13/19 (68%), Positives = 13/19 (68%), Gaps = 0/19 (0%)
Query 73 SGTNGCQFFITCGPCLWLD 91
S TNG QFFIT P WLD
Sbjct 504 SNTNGSQFFITVVPTPWLD 522
> SPAC57A10.03
Length=155
Score = 31.6 bits (70), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 20/49 (40%), Gaps = 17/49 (34%)
Query 43 HFRAGLLCMANSAAASSKQQQQQQQEQQQRSGTNGCQFFITCGPCLWLD 91
H AG+L MAN+ TN QFFIT P WLD
Sbjct 82 HTGAGILSMANAGP-----------------NTNSSQFFITLAPTPWLD 113
Lambda K H
0.317 0.125 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174483934
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40