bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1817_orf1
Length=112
Score E
Sequences producing significant alignments: (Bits) Value
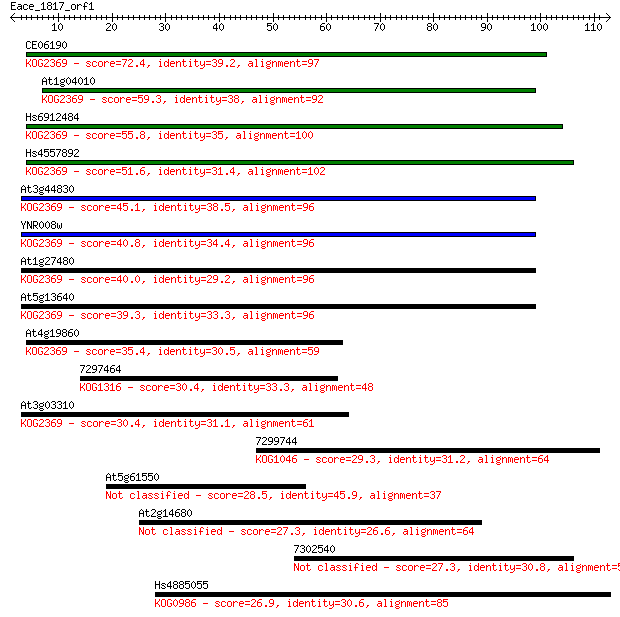
CE06190 72.4 2e-13
At1g04010 59.3 2e-09
Hs6912484 55.8 2e-08
Hs4557892 51.6 4e-07
At3g44830 45.1 3e-05
YNR008w 40.8 6e-04
At1g27480 40.0 0.001
At5g13640 39.3 0.002
At4g19860 35.4 0.026
7297464 30.4 0.89
At3g03310 30.4 0.94
7299744 29.3 2.1
At5g61550 28.5 3.4
At2g14680 27.3 6.5
7302540 27.3 6.7
Hs4885055 26.9 9.1
> CE06190
Length=417
Score = 72.4 bits (176), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 38/99 (38%), Positives = 60/99 (60%), Gaps = 2/99 (2%)
Query 4 GYQEGTSLIGIPYDWRLPPWQMD--FGRLKSEIESRVGAMGGAKADVLVHSLGSVVFNYF 61
GY+ G ++IG P+DWR P +++ +LKS IE+ K ++ HS+G+ + YF
Sbjct 146 GYRRGKNVIGAPFDWRKSPNELNDYLIQLKSLIETTYRWNDNQKIVLVGHSMGNPLSLYF 205
Query 62 LHKVVDQQWKDKYINSYTLVAPATGGSFKAIKALLTGYN 100
L+ VDQ WKDKYI+S+ +A GS + ++ +GYN
Sbjct 206 LNNYVDQAWKDKYISSFVSLAAPWAGSMQIVRLFASGYN 244
> At1g04010
Length=629
Score = 59.3 bits (142), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 35/104 (33%), Positives = 58/104 (55%), Gaps = 13/104 (12%)
Query 7 EGTSLIGIPYDWRLPPWQMD-----FGRLKSEIESRVGAMGGAKADVLVHSLGSVVFNYF 61
E +++ +PYDWRL P +++ F +LK E+ + GG V HS+G+ VF YF
Sbjct 146 EANAIVAVPYDWRLSPTKLEERDLYFHKLKLTFETALKLRGGPSI-VFAHSMGNNVFRYF 204
Query 62 LH----KVVDQ---QWKDKYINSYTLVAPATGGSFKAIKALLTG 98
L ++ + +W D++I++Y V GS +AIK+ L+G
Sbjct 205 LEWLRLEIAPKHYLKWLDQHIHAYFAVGAPLLGSVEAIKSTLSG 248
> Hs6912484
Length=412
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 35/102 (34%), Positives = 52/102 (50%), Gaps = 4/102 (3%)
Query 4 GYQEGTSLIGIPYDWRLPPWQMD--FGRLKSEIESRVGAMGGAKADVLVHSLGSVVFNYF 61
GY G + G PYDWR P + F L+ IE GG ++ HS+G++ YF
Sbjct 149 GYTRGEDVRGAPYDWRRAPNENGPYFLALREMIEEMYQLYGGPVV-LVAHSMGNMYTLYF 207
Query 62 LHKVVDQQWKDKYINSYTLVAPATGGSFKAIKALLTGYNDPV 103
L + Q WKDKYI ++ + GG K ++ L +G N+ +
Sbjct 208 LQRQ-PQAWKDKYIRAFVSLGAPWGGVAKTLRVLASGDNNRI 248
> Hs4557892
Length=440
Score = 51.6 bits (122), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 32/103 (31%), Positives = 52/103 (50%), Gaps = 2/103 (1%)
Query 4 GYQEGTSLIGIPYDWRLPPWQMD-FGRLKSEIESRVGAMGGAKADVLVHSLGSVVFNYFL 62
GY ++ PYDWRL P Q + + R + + + A G ++ HSLG + YFL
Sbjct 156 GYVRDETVRAAPYDWRLEPGQQEEYYRKLAGLVEEMHAAYGKPVFLIGHSLGCLHLLYFL 215
Query 63 HKVVDQQWKDKYINSYTLVAPATGGSFKAIKALLTGYNDPVDL 105
+ Q WKD++I+ + + GGS K + L +G N + +
Sbjct 216 LRQ-PQAWKDRFIDGFISLGAPWGGSIKPMLVLASGDNQGIPI 257
> At3g44830
Length=665
Score = 45.1 bits (105), Expect = 3e-05, Method: Composition-based stats.
Identities = 37/111 (33%), Positives = 50/111 (45%), Gaps = 16/111 (14%)
Query 3 MGYQEGTSLIGIPYDWRLPPWQMD-----FGRLKSEIESRVGAMGGAKADVLVHSLGSVV 57
+GY EG +L YDWRL + RLKS+IE G K V+ HS+G++
Sbjct 184 IGY-EGKNLHMASYDWRLSFHNTEVRDQSLSRLKSKIELMYATNGFKKVVVVPHSMGAIY 242
Query 58 FNYFLHKV----------VDQQWKDKYINSYTLVAPATGGSFKAIKALLTG 98
F +FL V W K+I S + PA G KA+ LL+
Sbjct 243 FLHFLKWVETPLPDGGGGGGPGWCAKHIKSVVNIGPAFLGVPKAVSNLLSA 293
> YNR008w
Length=661
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 33/111 (29%), Positives = 56/111 (50%), Gaps = 18/111 (16%)
Query 3 MGYQEGTSLIGIPYDWRLPPWQMD-----FGRLKSEIESRVGAMGGAKADVLVHSLGSVV 57
+GY E + YDWRL ++ F +LK +IE + G K ++ HS+GS +
Sbjct 272 IGY-EPNKMTSAAYDWRLAYLDLERRDRYFTKLKEQIE-LFHQLSGEKVCLIGHSMGSQI 329
Query 58 FNYFLHKVVDQQ----------WKDKYINSYTLVAPATGGSFKAIKALLTG 98
YF+ K V+ + W +++I+S+ A G+ KA+ AL++G
Sbjct 330 IFYFM-KWVEAEGPLYGNGGRGWVNEHIDSFINAAGTLLGAPKAVPALISG 379
> At1g27480
Length=413
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 28/105 (26%), Positives = 46/105 (43%), Gaps = 10/105 (9%)
Query 3 MGYQEGTSLIGIPYDWRL------PPWQMD---FGRLKSEIESRVGAMGGAKADVLVHSL 53
GY +++G PYD+R P ++ LK +E G +L HSL
Sbjct 132 CGYVNDQTILGAPYDFRYGLAASGHPSRVASQFLQDLKQLVEKTSSENEGKPVILLSHSL 191
Query 54 GSVVFNYFLHKVVDQQWKDKYINSYTLVAPATGGSFKAIKALLTG 98
G + +FL++ W+ KYI + +A GG+ +K +G
Sbjct 192 GGLFVLHFLNRTT-PSWRRKYIKHFVALAAPWGGTISQMKTFASG 235
> At5g13640
Length=671
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 32/110 (29%), Positives = 46/110 (41%), Gaps = 15/110 (13%)
Query 3 MGYQEGTSLIGIPYDWRLPPWQMD-----FGRLKSEIESRVGAMGGAKADVLVHSLGSVV 57
+GY+E + YDWRL + R+KS IE V GG KA ++ HS+G +
Sbjct 201 IGYEEKNMYMAA-YDWRLSFQNTEVRDQTLSRMKSNIELMVSTNGGKKAVIVPHSMGVLY 259
Query 58 FNYF---------LHKVVDQQWKDKYINSYTLVAPATGGSFKAIKALLTG 98
F +F L W KYI + + G KA+ L +
Sbjct 260 FLHFMKWVEAPAPLGGGGGPDWCAKYIKAVMNIGGPFLGVPKAVAGLFSA 309
> At4g19860
Length=493
Score = 35.4 bits (80), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 32/60 (53%), Gaps = 1/60 (1%)
Query 4 GYQEGTSLIGIPYDWRLPP-WQMDFGRLKSEIESRVGAMGGAKADVLVHSLGSVVFNYFL 62
G++EG +L G YD+R Q + ++E+ A G K +V+ HS+G ++ F+
Sbjct 133 GFEEGKTLFGFGYDFRQSNRLQETLDQFAKKLETVYKASGEKKINVISHSMGGLLVKCFM 192
> 7297464
Length=445
Score = 30.4 bits (67), Expect = 0.89, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 26/48 (54%), Gaps = 0/48 (0%)
Query 14 IPYDWRLPPWQMDFGRLKSEIESRVGAMGGAKADVLVHSLGSVVFNYF 61
+PYD RL +D + +E R G + A+AD LV +L + F++
Sbjct 31 LPYDSRLYADDLDASKAYAEALHRAGLINSAEADKLVKNLELLRFDWI 78
> At3g03310
Length=393
Score = 30.4 bits (67), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 35/62 (56%), Gaps = 1/62 (1%)
Query 3 MGYQEGTSLIGIPYDWRLPPW-QMDFGRLKSEIESRVGAMGGAKADVLVHSLGSVVFNYF 61
GY++GT+L G YD+R + LK ++E+ GG K ++ HS+G ++ + F
Sbjct 124 CGYKKGTTLFGYGYDFRQSNRIDLLILGLKKKLETAYKRSGGRKVTIISHSMGGLMVSCF 183
Query 62 LH 63
++
Sbjct 184 MY 185
> 7299744
Length=988
Score = 29.3 bits (64), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 20/67 (29%), Positives = 28/67 (41%), Gaps = 3/67 (4%)
Query 47 DVLVHSLGSVVFNY---FLHKVVDQQWKDKYINSYTLVAPATGGSFKAIKALLTGYNDPV 103
D + +S GS + FL + +Q Y+N Y TG F I L GYN
Sbjct 502 DTITYSKGSSLVRMLEDFLGETTFRQAVTNYLNEYKYSTAETGNFFTEIDKLELGYNVTE 561
Query 104 DLTFWTL 110
+ WT+
Sbjct 562 IMLTWTV 568
> At5g61550
Length=845
Score = 28.5 bits (62), Expect = 3.4, Method: Composition-based stats.
Identities = 17/37 (45%), Positives = 19/37 (51%), Gaps = 4/37 (10%)
Query 19 RLPPWQMDFGRLKSEIESRVGAMGGAKADVLVHSLGS 55
RLPP DF SE +SRV A L HS+GS
Sbjct 206 RLPPEYQDFLSAVSEAQSRVSPFSPA----LKHSMGS 238
> At2g14680
Length=629
Score = 27.3 bits (59), Expect = 6.5, Method: Composition-based stats.
Identities = 17/65 (26%), Positives = 31/65 (47%), Gaps = 3/65 (4%)
Query 25 MDFGRLKSEIESRVGAMGGAKADVLVHSLGSVVFNYFLHKVVDQQWKDKYINSYTLVAPA 84
M+ +L+ E +S M G + + + G +V + + WK YIN + + P+
Sbjct 435 MELAQLRQEYQSLRDKMSGTREEESTGNSGRIVISNEKDGRLKNMWKKSYINRW--IDPS 492
Query 85 T-GGS 88
+ GGS
Sbjct 493 SRGGS 497
> 7302540
Length=474
Score = 27.3 bits (59), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 16/52 (30%), Positives = 22/52 (42%), Gaps = 0/52 (0%)
Query 54 GSVVFNYFLHKVVDQQWKDKYINSYTLVAPATGGSFKAIKALLTGYNDPVDL 105
G F + + +D Q KDKY N GG F A++ Y D +D
Sbjct 410 GCARFKSDIMQFLDVQLKDKYTNLVHSTYHKKGGHFAALEVPKVLYKDFIDF 461
> Hs4885055
Length=688
Score = 26.9 bits (58), Expect = 9.1, Method: Composition-based stats.
Identities = 26/87 (29%), Positives = 34/87 (39%), Gaps = 15/87 (17%)
Query 28 GRLKSEIESRVGAMGGAKADVLVHSLGSVVFNYFLHKVVDQQWKDKYINSY--TLVAPAT 85
G L+ ++ R+G GG +V HS K VD W+ Y+ Y L+ P
Sbjct 427 GLLQRDVSKRLGCHGGGSQEVKEHS---------FFKGVD--WQHVYLQKYPPPLIPPR- 474
Query 86 GGSFKAIKALLTGYNDPVDLTFWTLLD 112
G A A G D D LLD
Sbjct 475 -GEVNAADAFDIGSFDEEDTKGIKLLD 500
Lambda K H
0.321 0.139 0.453
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1188972946
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40