bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1744_orf1
Length=154
Score E
Sequences producing significant alignments: (Bits) Value
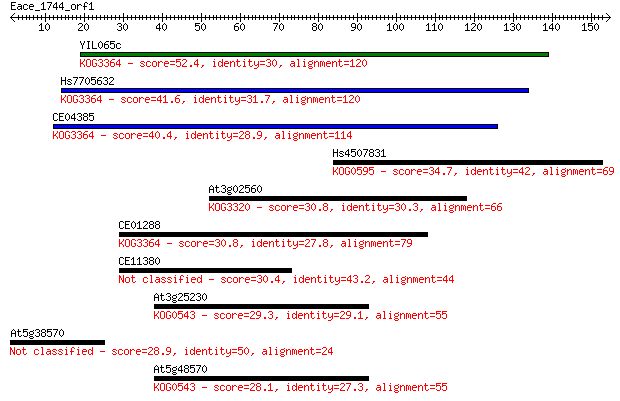
YIL065c 52.4 4e-07
Hs7705632 41.6 6e-04
CE04385 40.4 0.002
Hs4507831 34.7 0.076
At3g02560 30.8 0.97
CE01288 30.8 1.0
CE11380 30.4 1.6
At3g25230 29.3 3.2
At5g38570 28.9 4.0
At5g48570 28.1 6.6
> YIL065c
Length=155
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 36/123 (29%), Positives = 59/123 (47%), Gaps = 8/123 (6%)
Query 19 LESGCPCPRLQ--FEFASLLICSPNVKDLKDSADLLTELL-EIGFCRSDCLFQLALVYLK 75
+ G P +Q F +A LI S +V D + +LT++ E R +CL+ L + K
Sbjct 30 VSEGGPTATIQSRFNYAWGLIKSTDVNDERLGVKILTDIYKEAESRRRECLYYLTIGCYK 89
Query 76 LGHYSLAKRRVEALLRMGLGVQEPRNLSALSLHSLILDRAAYDGVSGSLILGLIAGGVLP 135
LG YS+AKR V+ L E N +L S++ D+ + + G ++ G + G +
Sbjct 90 LGEYSMAKRYVDTLFE-----HERNNKQVGALKSMVEDKIQKETLKGVVVAGGVLAGAVA 144
Query 136 SGS 138
S
Sbjct 145 VAS 147
> Hs7705632
Length=152
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 38/122 (31%), Positives = 55/122 (45%), Gaps = 7/122 (5%)
Query 14 KYLEQLESGCPCPRLQFEFASLLICSPNVKDLKDSADLLTELLEIGFC--RSDCLFQLAL 71
K+ + +G QFE+A L+ + D++ LL ELL G + D +F
Sbjct 20 KFQSEKAAGSVSKSTQFEYAWCLVRTRYNDDIRKGIVLLEELLPKGSKEEQRDYVF---- 75
Query 72 VYLKLGHYSLAKRRVEALLRMGLGVQEPRNLSALSLHSLILDRAAYDGVSGSLILGLIAG 131
YL +G+Y L + GL EP+N A L LI DG+ G I+G +A
Sbjct 76 -YLAVGNYRLKEYEKALKYVRGLLQTEPQNNQAKELERLIDKAMKKDGLVGMAIVGGMAL 134
Query 132 GV 133
GV
Sbjct 135 GV 136
> CE04385
Length=138
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 33/117 (28%), Positives = 53/117 (45%), Gaps = 8/117 (6%)
Query 12 RAKYLEQLESGCPCPRLQFEFASLLICSPNVKDLKDSADLLTELL---EIGFCRSDCLFQ 68
R +Y+ Q G P F FA +I S N D+K+ L +LL E + + ++
Sbjct 7 REQYMRQCARGDPSAASTFAFAHAMIGSKNKLDVKEGIVCLEKLLRDDEDRTSKRNYVYY 66
Query 69 LALVYLKLGHYSLAKRRVEALLRMGLGVQEPRNLSALSLHSLILDRAAYDGVSGSLI 125
LA+ + ++ Y LA ++ LL E N A +L I +DG+ G+ I
Sbjct 67 LAVAHARIKQYDLALGYIDVLLDA-----EGDNQQAKTLKESIKSAMTHDGLIGAAI 118
> Hs4507831
Length=1050
Score = 34.7 bits (78), Expect = 0.076, Method: Compositional matrix adjust.
Identities = 29/82 (35%), Positives = 33/82 (40%), Gaps = 13/82 (15%)
Query 84 RRVEALLRMGLGVQEPRNLSALSLHSLILDRAAYDGVSGSLILGLIAGGV---------- 133
R + LL+ G Q P S SL ++ A G GSL G AGG
Sbjct 697 RLTDLLLKAAFGTQAPDPGSTESLQEKPMEIAPSAGFGGSLHPGARAGGTSSPSPVVFTV 756
Query 134 --LPSGS-PPQQVRWRSLDRGP 152
PSGS PPQ R R GP
Sbjct 757 GSPPSGSTPPQGPRTRMFSAGP 778
> At3g02560
Length=191
Score = 30.8 bits (68), Expect = 0.97, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 28/66 (42%), Gaps = 13/66 (19%)
Query 52 LTELLEIGFCRSDCLFQLALVYLKLGHYSLAKRRVEALLRMGLGVQEPRNLSALSLHSLI 111
L LE F D +F +A RR+ + G VQ PRN + S+H +
Sbjct 80 LVRELEKKFSGKDVIF-------------VATRRIMRPPKKGSAVQRPRNRTLTSVHEAM 126
Query 112 LDRAAY 117
L+ AY
Sbjct 127 LEDVAY 132
> CE01288
Length=143
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 22/82 (26%), Positives = 41/82 (50%), Gaps = 8/82 (9%)
Query 29 QFEFASLLICSPNVKDLKDSADLLTELLEIGFCRSD---CLFQLALVYLKLGHYSLAKRR 85
Q A +L+ S + +++K+ ++L +++ D C+ LAL + +L +Y +
Sbjct 29 QISLAIVLVGSEDRREIKEGIEILEDVVSDTAHSEDSRVCVHYLALAHARLKNYDKSINL 88
Query 86 VEALLRMGLGVQEPRNLSALSL 107
+ ALLR EP N+ A L
Sbjct 89 LNALLRT-----EPSNMQATEL 105
> CE11380
Length=276
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 19/44 (43%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 29 QFEFASLLICSPNVKDLKDSADLLTELLEIGFCRSDCLFQLALV 72
Q FA LLI S K+ KD LLT+ L+ DC L LV
Sbjct 22 QMSFAELLIISILSKNAKDFVTLLTKPLKNMIISVDCWLSLKLV 65
> At3g25230
Length=551
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 16/55 (29%), Positives = 28/55 (50%), Gaps = 0/55 (0%)
Query 38 CSPNVKDLKDSADLLTELLEIGFCRSDCLFQLALVYLKLGHYSLAKRRVEALLRM 92
C +KD K + L T++LE+ L++ A Y++L LA+ V+ L +
Sbjct 458 CKLKLKDYKQAEKLCTKVLELESTNVKALYRRAQAYMELSDLDLAEFDVKKALEI 512
> At5g38570
Length=411
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 12/24 (50%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 1 SFDLKAETEALRAKYLEQLESGCP 24
+ L+AE+ + KYL+QL SGCP
Sbjct 116 TLQLEAESYFIDGKYLQQLISGCP 139
> At5g48570
Length=578
Score = 28.1 bits (61), Expect = 6.6, Method: Composition-based stats.
Identities = 15/55 (27%), Positives = 29/55 (52%), Gaps = 0/55 (0%)
Query 38 CSPNVKDLKDSADLLTELLEIGFCRSDCLFQLALVYLKLGHYSLAKRRVEALLRM 92
C +KD K++A L T++LE+ +++ A YL+ LA+ ++ L +
Sbjct 468 CKLKLKDYKEAAKLSTKVLEMDSRNVKAMYRRAHAYLETADLDLAELDIKKALEI 522
Lambda K H
0.322 0.140 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1997677330
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40