bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1705_orf1
Length=177
Score E
Sequences producing significant alignments: (Bits) Value
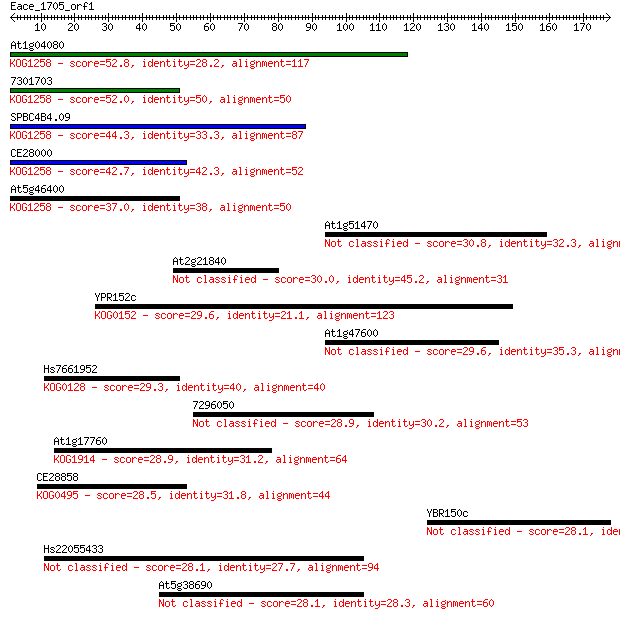
At1g04080 52.8 3e-07
7301703 52.0 6e-07
SPBC4B4.09 44.3 1e-04
CE28000 42.7 3e-04
At5g46400 37.0 0.020
At1g51470 30.8 1.3
At2g21840 30.0 2.5
YPR152c 29.6 2.9
At1g47600 29.6 3.4
Hs7661952 29.3 4.2
7296050 28.9 5.7
At1g17760 28.9 6.3
CE28858 28.5 7.5
YBR150c 28.1 8.5
Hs22055433 28.1 9.3
At5g38690 28.1 9.4
> At1g04080
Length=768
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 33/122 (27%), Positives = 62/122 (50%), Gaps = 5/122 (4%)
Query 1 FWHPDPLAPRRLQAWRDYLDFEEKNAP-ERLPLLQRRCLEVCASYPELWLRCALQKKKSS 59
++H PL L+ W +YLDF E++ ++ L RC+ CA+YPE W+R + S
Sbjct 350 YFHVRPLNVAELENWHNYLDFIERDGDFNKVVKLYERCVVTCANYPEYWIRYVTNMEASG 409
Query 60 EEEALQLLHFAATKVLKRRRDMACIYASQL-EACGRLKEAAKEFEAL---VRPPLDSASL 115
+ + AT+V +++ ++A++L E G + A ++ + + P L A +
Sbjct 410 SADLAENALARATQVFVKKQPEIHLFAARLKEQNGDIAGARAAYQLVHSEISPGLLEAVI 469
Query 116 KY 117
K+
Sbjct 470 KH 471
> 7301703
Length=1009
Score = 52.0 bits (123), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 25/51 (49%), Positives = 33/51 (64%), Gaps = 1/51 (1%)
Query 1 FWHPDPLAPRRLQAWRDYLDFE-EKNAPERLPLLQRRCLEVCASYPELWLR 50
++H PL +L+ W+DYLDFE EK ER+ +L RCL CA Y E WL+
Sbjct 637 YFHVKPLERAQLKNWKDYLDFEIEKGDRERVLVLFERCLIACALYDEFWLK 687
> SPBC4B4.09
Length=612
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/88 (32%), Positives = 39/88 (44%), Gaps = 10/88 (11%)
Query 1 FWHPDPLAPRRLQAWRDYLDFEEKNAP-ERLPLLQRRCLEVCASYPELWLRCALQKKKSS 59
++H L +L WR YLDFEE +R+ L RCL CA Y E W R A
Sbjct 271 YFHVKELDEAQLVNWRKYLDFEEVEGDFQRICHLYERCLITCALYDEFWFRYARWMSAQP 330
Query 60 EEEALQLLHFAATKVLKRRRDMACIYAS 87
+ H ++ R +CI+AS
Sbjct 331 D-------HLNDVSIIYER--ASCIFAS 349
> CE28000
Length=710
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/53 (41%), Positives = 30/53 (56%), Gaps = 1/53 (1%)
Query 1 FWHPDPLAPRRLQAWRDYLDFEEKNA-PERLPLLQRRCLEVCASYPELWLRCA 52
++H PL +L W YLDFE K ER+ +L RCL C+ Y E W++ A
Sbjct 373 YFHVKPLDYPQLFNWMSYLDFEIKEGHEERVKILFDRCLIPCSLYEEFWIKYA 425
> At5g46400
Length=1022
Score = 37.0 bits (84), Expect = 0.020, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 24/51 (47%), Gaps = 1/51 (1%)
Query 1 FWHPDPLAPRRLQAWRDYLDFEEKNAPERLPL-LQRRCLEVCASYPELWLR 50
++H PL +L W YL F E + L RCL CA+Y E W R
Sbjct 287 YFHVKPLDTNQLDNWHAYLSFGETYGDFDWAINLYERCLIPCANYTEFWFR 337
> At1g51470
Length=465
Score = 30.8 bits (68), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 31/65 (47%), Gaps = 6/65 (9%)
Query 94 RLKEAAKEFEALVRPPLDSASLKYFVALLQFSLRHPPETAADPLSHALLLLEEAADKYRD 153
RL E E ALV+ LD L Y+V+ Q++ PP T + ++ A + L YR+
Sbjct 282 RLPEFTPEESALVKGSLDFLGLNYYVS--QYATDAPPPTQPNAITDARVTL----GFYRN 335
Query 154 DGPCA 158
P
Sbjct 336 GSPIG 340
> At2g21840
Length=746
Score = 30.0 bits (66), Expect = 2.5, Method: Composition-based stats.
Identities = 14/31 (45%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 49 LRCALQKKKSSEEEALQLLHFAATKVLKRRR 79
L CALQK ++ EA ++LH++ +LKR R
Sbjct 106 LGCALQKNIATSWEAKEMLHYSHEHLLKRCR 136
> YPR152c
Length=465
Score = 29.6 bits (65), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 26/131 (19%), Positives = 61/131 (46%), Gaps = 10/131 (7%)
Query 26 APERLPLLQRRCLEVCASYPELWLRCALQKKKSSEEEALQLLHFAATKVLKRRRDMACI- 84
AP+ +P R+ + A Y ++ + K+ ++ QLL + T L++R+++ C
Sbjct 308 APDTIPQDIRK--QQKALYKAYKIKEYIPSKRDQDKFVSQLLFYYKTFDLEQRKEIFCDC 365
Query 85 -------YASQLEACGRLKEAAKEFEALVRPPLDSASLKYFVALLQFSLRHPPETAADPL 137
+ +E+ + KE ++ L++ P DS+S++ + ++ P +P
Sbjct 366 LRDHERDFTGAVESLRQDKELIDRWQTLLKAPADSSSIEDILLSIEHRCCVSPIVVTEPR 425
Query 138 SHALLLLEEAA 148
+ + +LE+
Sbjct 426 YYVVGILEKTV 436
> At1g47600
Length=510
Score = 29.6 bits (65), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 26/51 (50%), Gaps = 2/51 (3%)
Query 94 RLKEAAKEFEALVRPPLDSASLKYFVALLQFSLRHPPETAADPLSHALLLL 144
RL E E ALV+ LD L Y+V Q++ PP T + ++ A + L
Sbjct 327 RLPEFTPEQSALVKGSLDFLGLNYYVT--QYATDAPPPTQLNAITDARVTL 375
> Hs7661952
Length=963
Score = 29.3 bits (64), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 23/41 (56%), Gaps = 1/41 (2%)
Query 11 RLQAWRDYLDFEEK-NAPERLPLLQRRCLEVCASYPELWLR 50
RL ++ Y+DFE K P R+ L+ R L P+LW+R
Sbjct 309 RLAEYQAYIDFEMKIGDPARIQLIFERALVENCLVPDLWIR 349
> 7296050
Length=951
Score = 28.9 bits (63), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 28/53 (52%), Gaps = 0/53 (0%)
Query 55 KKKSSEEEALQLLHFAATKVLKRRRDMACIYASQLEACGRLKEAAKEFEALVR 107
+++ E+E L A +RR +A + S ++ GRLK+ K+ E L+R
Sbjct 31 RQRDKEQEQQDLACLAKRDYTERRNGLAVLKNSGRKSTGRLKDNLKKLEDLLR 83
> At1g17760
Length=722
Score = 28.9 bits (63), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 20/72 (27%), Positives = 31/72 (43%), Gaps = 15/72 (20%)
Query 14 AWRDYLDFEEKN--------APERLPLLQRRCLEVCASYPELWLRCALQKKKSSEEEALQ 65
AW+ +L FE+ N + +R+ +CL YP++W A KS +
Sbjct 227 AWKKFLSFEKGNPQRIDTASSTKRIIYAYEQCLMCLYHYPDVWYDYAEWHVKSGSTD--- 283
Query 66 LLHFAATKVLKR 77
AA KV +R
Sbjct 284 ----AAIKVFQR 291
> CE28858
Length=968
Score = 28.5 bits (62), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 14/44 (31%), Positives = 24/44 (54%), Gaps = 3/44 (6%)
Query 9 PRRLQAWRDYLDFEEKNAPERLPLLQRRCLEVCASYPELWLRCA 52
P ++ W+ ++ E+ PE +L R +E C+S E+WL A
Sbjct 427 PSSVKLWKAAVELED---PEDARILLTRAVECCSSSTEMWLALA 467
> YBR150c
Length=1094
Score = 28.1 bits (61), Expect = 8.5, Method: Composition-based stats.
Identities = 16/54 (29%), Positives = 26/54 (48%), Gaps = 0/54 (0%)
Query 124 FSLRHPPETAADPLSHALLLLEEAADKYRDDGPCAEVLHNYRAKLVALHAGDTK 177
FS+R + D + +L ++A D+ RDD P L NY+ + L +K
Sbjct 621 FSIRTTLDDTLDEIIENVLENQKALDRMRDDLPTILSLENYKENMRILSLDSSK 674
> Hs22055433
Length=1416
Score = 28.1 bits (61), Expect = 9.3, Method: Composition-based stats.
Identities = 26/97 (26%), Positives = 50/97 (51%), Gaps = 11/97 (11%)
Query 11 RLQAWRDYLDFEEKNAPERLPLLQRRCLEVCASYPELWLRCALQKKKSSEEEALQLLHFA 70
+ +A + L+ + + E+L L R+ EVC +KK+ S ++ +LLHF+
Sbjct 1228 KYKATKSDLETQISSLNEKLANLNRKYEEVCEEV------LHAKKKEISAKDEKELLHFS 1281
Query 71 ATKVL---KRRRDMACIYASQLEACGRLKEAAKEFEA 104
+ + K R D + ++L+ R++E+AK+ EA
Sbjct 1282 IEQEIKDQKERCDKSLTTITELQR--RIQESAKQIEA 1316
> At5g38690
Length=544
Score = 28.1 bits (61), Expect = 9.4, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 31/60 (51%), Gaps = 0/60 (0%)
Query 45 PELWLRCALQKKKSSEEEALQLLHFAATKVLKRRRDMACIYASQLEACGRLKEAAKEFEA 104
PE++ + Q +K + + L+LL+F + L CI + +E+ R KEA ++ A
Sbjct 354 PEMFEKGISQYEKLNSSKRLKLLNFLCDETLGTLVMRNCIDSQNIESVERKKEAKEKINA 413
Lambda K H
0.322 0.135 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2743263016
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40