bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1693_orf2
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
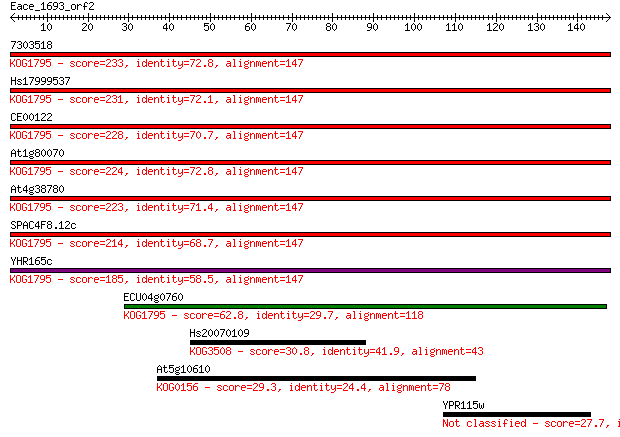
7303518 233 9e-62
Hs17999537 231 3e-61
CE00122 228 3e-60
At1g80070 224 5e-59
At4g38780 223 1e-58
SPAC4F8.12c 214 7e-56
YHR165c 185 2e-47
ECU04g0760 62.8 3e-10
Hs20070109 30.8 0.85
At5g10610 29.3 2.6
YPR115w 27.7 8.3
> 7303518
Length=2396
Score = 233 bits (594), Expect = 9e-62, Method: Composition-based stats.
Identities = 107/147 (72%), Positives = 122/147 (82%), Gaps = 0/147 (0%)
Query 1 WEISNRLPRSLTTLNWSSSFASVYSLDNPNLLFSMGGFECRILPKVRMQTEEFSQREGAW 60
W+I NRLPRS+TT+ W S+F SVYS DNPNLLF+M GFECRILPK R Q EEF+ R+G W
Sbjct 1215 WDIKNRLPRSVTTIGWESTFVSVYSKDNPNLLFNMSGFECRILPKCRTQNEEFTHRDGVW 1274
Query 61 KLQNETTKELAAQAFLKVGDEGMRRFENRVRMVLMSSGATTFTKIANKWNTTLISLMTYF 120
LQNE TKE AQ FL+V DE + RF NRVR +LM+SG+TTFTKI NKWNT LI LMTYF
Sbjct 1275 NLQNEITKERTAQCFLRVDDESLGRFHNRVRQILMASGSTTFTKIVNKWNTALIGLMTYF 1334
Query 121 REAVIHTEALLDLLVKCENKIQTRIKI 147
REAV++T+ LLDLLVKCENKIQTRIKI
Sbjct 1335 REAVVNTQELLDLLVKCENKIQTRIKI 1361
> Hs17999537
Length=2335
Score = 231 bits (590), Expect = 3e-61, Method: Compositional matrix adjust.
Identities = 106/147 (72%), Positives = 122/147 (82%), Gaps = 0/147 (0%)
Query 1 WEISNRLPRSLTTLNWSSSFASVYSLDNPNLLFSMGGFECRILPKVRMQTEEFSQREGAW 60
W+I NRLPRS+TT+ W +SF SVYS DNPNLLF+M GFECRILPK R EEF+ ++G W
Sbjct 1155 WDIKNRLPRSVTTVQWENSFVSVYSKDNPNLLFNMCGFECRILPKCRTSYEEFTHKDGVW 1214
Query 61 KLQNETTKELAAQAFLKVGDEGMRRFENRVRMVLMSSGATTFTKIANKWNTTLISLMTYF 120
LQNE TKE AQ FL+V DE M+RF NRVR +LM+SG+TTFTKI NKWNT LI LMTYF
Sbjct 1215 NLQNEVTKERTAQCFLRVDDESMQRFHNRVRQILMASGSTTFTKIVNKWNTALIGLMTYF 1274
Query 121 REAVIHTEALLDLLVKCENKIQTRIKI 147
REAV++T+ LLDLLVKCENKIQTRIKI
Sbjct 1275 REAVVNTQELLDLLVKCENKIQTRIKI 1301
> CE00122
Length=2329
Score = 228 bits (581), Expect = 3e-60, Method: Compositional matrix adjust.
Identities = 104/147 (70%), Positives = 119/147 (80%), Gaps = 0/147 (0%)
Query 1 WEISNRLPRSLTTLNWSSSFASVYSLDNPNLLFSMGGFECRILPKVRMQTEEFSQREGAW 60
W+I NRLPRS+TT+ W +SF SVYS DNPN+LF M GFECRILPK R EEF R+G W
Sbjct 1147 WDIKNRLPRSITTVEWENSFVSVYSKDNPNMLFDMSGFECRILPKCRTANEEFVHRDGVW 1206
Query 61 KLQNETTKELAAQAFLKVGDEGMRRFENRVRMVLMSSGATTFTKIANKWNTTLISLMTYF 120
LQNE TKE AQ FLKV +E + +F NR+R +LMSSG+TTFTKI NKWNT LI LMTYF
Sbjct 1207 NLQNEVTKERTAQCFLKVDEESLSKFHNRIRQILMSSGSTTFTKIVNKWNTALIGLMTYF 1266
Query 121 REAVIHTEALLDLLVKCENKIQTRIKI 147
REAV++T+ LLDLLVKCENKIQTRIKI
Sbjct 1267 REAVVNTQELLDLLVKCENKIQTRIKI 1293
> At1g80070
Length=2382
Score = 224 bits (571), Expect = 5e-59, Method: Compositional matrix adjust.
Identities = 107/148 (72%), Positives = 120/148 (81%), Gaps = 1/148 (0%)
Query 1 WEISNRLPRSLTTLNWSSSFASVYSLDNPNLLFSMGGFECRILPKVRMQTEEFSQ-REGA 59
W++ NRLPRS+TTL W + F SVYS DNPNLLFSM GFE RILPK+RM E FS ++G
Sbjct 1201 WDMKNRLPRSITTLEWENGFVSVYSKDNPNLLFSMCGFEVRILPKIRMTQEAFSNTKDGV 1260
Query 60 WKLQNETTKELAAQAFLKVGDEGMRRFENRVRMVLMSSGATTFTKIANKWNTTLISLMTY 119
W LQNE TKE A AFL+V DE M+ FENRVR +LMSSG+TTFTKI NKWNT LI LMTY
Sbjct 1261 WNLQNEQTKERTAVAFLRVDDEHMKVFENRVRQILMSSGSTTFTKIVNKWNTALIGLMTY 1320
Query 120 FREAVIHTEALLDLLVKCENKIQTRIKI 147
FREA +HT+ LLDLLVKCENKIQTRIKI
Sbjct 1321 FREATVHTQELLDLLVKCENKIQTRIKI 1348
> At4g38780
Length=2352
Score = 223 bits (567), Expect = 1e-58, Method: Compositional matrix adjust.
Identities = 105/148 (70%), Positives = 119/148 (80%), Gaps = 1/148 (0%)
Query 1 WEISNRLPRSLTTLNWSSSFASVYSLDNPNLLFSMGGFECRILPKVRMQTEEFSQ-REGA 59
W++ NRLPRS+TTL W + F SVYS DNPNLLFSM GFE R+LPK+RM E FS R+G
Sbjct 1173 WDMKNRLPRSITTLEWENGFVSVYSKDNPNLLFSMCGFEVRVLPKIRMGQEAFSSTRDGV 1232
Query 60 WKLQNETTKELAAQAFLKVGDEGMRRFENRVRMVLMSSGATTFTKIANKWNTTLISLMTY 119
W LQNE TKE A AFL+ DE M+ FENRVR +LMSSG+TTFTKI NKWNT LI LMTY
Sbjct 1233 WNLQNEQTKERTAVAFLRADDEHMKVFENRVRQILMSSGSTTFTKIVNKWNTALIGLMTY 1292
Query 120 FREAVIHTEALLDLLVKCENKIQTRIKI 147
FREA +HT+ LLDLLVKCENKIQTR+KI
Sbjct 1293 FREATVHTQELLDLLVKCENKIQTRVKI 1320
> SPAC4F8.12c
Length=2363
Score = 214 bits (544), Expect = 7e-56, Method: Compositional matrix adjust.
Identities = 101/147 (68%), Positives = 120/147 (81%), Gaps = 1/147 (0%)
Query 1 WEISNRLPRSLTTLNWSSSFASVYSLDNPNLLFSMGGFECRILPKVRMQTEEFSQREGAW 60
WEI NRLPRSLTTL W +F SVYS DNPNLLFSM GFE RILPK+R Q EEFS ++G W
Sbjct 1180 WEIRNRLPRSLTTLEWEDTFPSVYSKDNPNLLFSMTGFEVRILPKIR-QNEEFSLKDGVW 1238
Query 61 KLQNETTKELAAQAFLKVGDEGMRRFENRVRMVLMSSGATTFTKIANKWNTTLISLMTYF 120
L + TK+ AQAF++V ++G+ +F NR+R +LMSSG+TTFTKIANKWNT LI+LMTY+
Sbjct 1239 NLTDNRTKQRTAQAFIRVTEDGINQFGNRIRQILMSSGSTTFTKIANKWNTALIALMTYY 1298
Query 121 REAVIHTEALLDLLVKCENKIQTRIKI 147
REA I T LLDLLVKCE+KIQTR+KI
Sbjct 1299 REAAISTPELLDLLVKCESKIQTRVKI 1325
> YHR165c
Length=2413
Score = 185 bits (470), Expect = 2e-47, Method: Compositional matrix adjust.
Identities = 86/147 (58%), Positives = 117/147 (79%), Gaps = 1/147 (0%)
Query 1 WEISNRLPRSLTTLNWSSSFASVYSLDNPNLLFSMGGFECRILPKVRMQTEEFSQREGAW 60
WEI +R+P SLT++ W ++F SVYS +NPNLLFSM GFE RILP+ RM+ E S EG W
Sbjct 1228 WEIQSRVPTSLTSIKWENAFVSVYSKNNPNLLFSMCGFEVRILPRQRME-EVVSNDEGVW 1286
Query 61 KLQNETTKELAAQAFLKVGDEGMRRFENRVRMVLMSSGATTFTKIANKWNTTLISLMTYF 120
L +E TK+ A+A+LKV +E +++F++R+R +LM+SG+TTFTK+A KWNT+LISL TYF
Sbjct 1287 DLVDERTKQRTAKAYLKVSEEEIKKFDSRIRGILMASGSTTFTKVAAKWNTSLISLFTYF 1346
Query 121 REAVIHTEALLDLLVKCENKIQTRIKI 147
REA++ TE LLD+LVK E +IQ R+K+
Sbjct 1347 REAIVATEPLLDILVKGETRIQNRVKL 1373
> ECU04g0760
Length=2172
Score = 62.8 bits (151), Expect = 3e-10, Method: Composition-based stats.
Identities = 35/118 (29%), Positives = 65/118 (55%), Gaps = 13/118 (11%)
Query 29 PNLLFSMGGFECRILPKVRMQTEEFSQREGAWKLQNETTKELAAQAFLKVGDEGMRRFEN 88
P + FSM G + V + +++ +R +W+L N A L V + G+ FE+
Sbjct 1192 PFVKFSMVGVD------VLISSKKIHERS-SWRLGN------GMHANLAVSEGGIEMFES 1238
Query 89 RVRMVLMSSGATTFTKIANKWNTTLISLMTYFREAVIHTEALLDLLVKCENKIQTRIK 146
+ ++ +SG+ TF K+A +WNT +++ +TY+RE + T+ L++ L + E I +K
Sbjct 1239 NISHIVSTSGSATFLKVATRWNTQILAFVTYYRECICDTKGLVEKLQRAERLIGNVVK 1296
> Hs20070109
Length=967
Score = 30.8 bits (68), Expect = 0.85, Method: Compositional matrix adjust.
Identities = 18/45 (40%), Positives = 29/45 (64%), Gaps = 2/45 (4%)
Query 45 KVRMQTEEFSQREGAWKLQNETTKELA--AQAFLKVGDEGMRRFE 87
K+R+ T++ + E +K Q ETT++L QA L+ G+E +RR E
Sbjct 855 KLRISTKKLEEYETLFKCQEETTQKLVLEYQARLEEGEERLRRHE 899
> At5g10610
Length=500
Score = 29.3 bits (64), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 19/81 (23%), Positives = 39/81 (48%), Gaps = 3/81 (3%)
Query 37 GFECRILPKVRMQTEEFSQREGAWKLQNETTKELAAQAFLKVGDEGMRRFENRVR---MV 93
G E R++ RM+ E + +++N + + FLK+ + + + V +V
Sbjct 235 GLEKRVIDMQRMRDEYLQRLIDDIRMKNIDSSGSVVEKFLKLQESEPEFYADDVIKGIIV 294
Query 94 LMSSGATTFTKIANKWNTTLI 114
LM +G T + +A +W +L+
Sbjct 295 LMFNGGTDTSPVAMEWAVSLL 315
> YPR115w
Length=1083
Score = 27.7 bits (60), Expect = 8.3, Method: Composition-based stats.
Identities = 12/36 (33%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 107 NKWNTTLISLMTYFREAVIHTEALLDLLVKCENKIQ 142
+KW L SL+ YFREA E + + + +N ++
Sbjct 164 HKWKKILKSLIAYFREAAYSQEQIARINYQMKNAVK 199
Lambda K H
0.320 0.131 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1785281974
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40