bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1650_orf1
Length=98
Score E
Sequences producing significant alignments: (Bits) Value
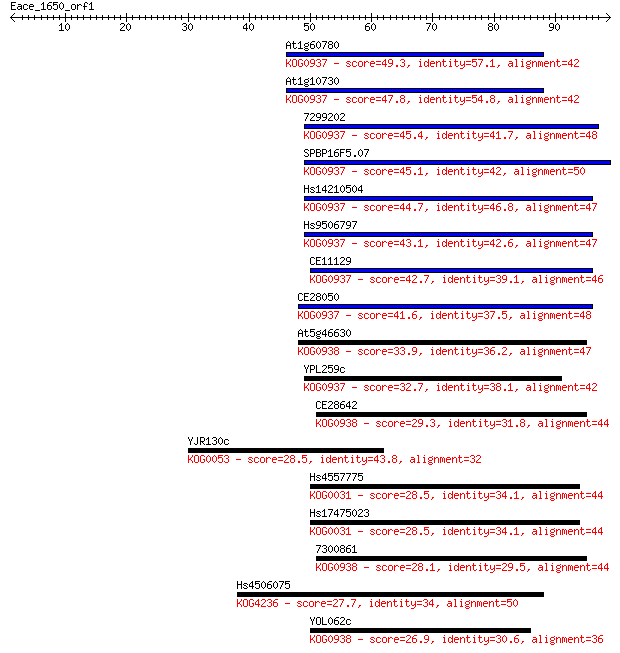
At1g60780 49.3 2e-06
At1g10730 47.8 5e-06
7299202 45.4 3e-05
SPBP16F5.07 45.1 4e-05
Hs14210504 44.7 5e-05
Hs9506797 43.1 1e-04
CE11129 42.7 2e-04
CE28050 41.6 4e-04
At5g46630 33.9 0.069
YPL259c 32.7 0.15
CE28642 29.3 2.0
YJR130c 28.5 3.3
Hs4557775 28.5 3.4
Hs17475023 28.5 3.5
7300861 28.1 4.7
Hs4506075 27.7 5.0
YOL062c 26.9 9.7
> At1g60780
Length=428
Score = 49.3 bits (116), Expect = 2e-06, Method: Composition-based stats.
Identities = 24/43 (55%), Positives = 35/43 (81%), Gaps = 1/43 (2%)
Query 46 MAGA-SAVLVLDGRGKVIIGRDYRGDVSLAISERFACRVIDQE 87
MAGA SA+ +LD +G+V++ RDYRGDVS A +ERF ++I++E
Sbjct 1 MAGAASALFLLDIKGRVLVWRDYRGDVSAAQAERFFTKLIEKE 43
> At1g10730
Length=428
Score = 47.8 bits (112), Expect = 5e-06, Method: Composition-based stats.
Identities = 23/43 (53%), Positives = 34/43 (79%), Gaps = 1/43 (2%)
Query 46 MAGA-SAVLVLDGRGKVIIGRDYRGDVSLAISERFACRVIDQE 87
MAGA SA+ +LD +G+V++ RDYRGDV+ A +ERF ++I+ E
Sbjct 1 MAGAASALFLLDIKGRVLVWRDYRGDVTAAQAERFFTKLIETE 43
> 7299202
Length=426
Score = 45.4 bits (106), Expect = 3e-05, Method: Composition-based stats.
Identities = 20/50 (40%), Positives = 37/50 (74%), Gaps = 2/50 (4%)
Query 49 ASAVLVLDGRGKVIIGRDYRGD-VSLAISERFACRVID-QEEAAVTPVFE 96
+SA+ VLD +GKV+I R+YRGD + +A+ ++F +++ +EE +TP+ +
Sbjct 3 SSAIFVLDVKGKVLISRNYRGDNIDMAVIDKFMPLLMEREEEGLITPILQ 52
> SPBP16F5.07
Length=426
Score = 45.1 bits (105), Expect = 4e-05, Method: Composition-based stats.
Identities = 21/52 (40%), Positives = 33/52 (63%), Gaps = 2/52 (3%)
Query 49 ASAVLVLDGRGKVIIGRDYRGDVSLAISERFAC--RVIDQEEAAVTPVFEDE 98
ASA+ VL+ +GKVII RDYR D+ +++ E+F +++E+ TP E
Sbjct 2 ASAIFVLNLKGKVIISRDYRADIPMSVVEKFLPLKSEVEEEQGFSTPCLTHE 53
> Hs14210504
Length=423
Score = 44.7 bits (104), Expect = 5e-05, Method: Composition-based stats.
Identities = 22/48 (45%), Positives = 34/48 (70%), Gaps = 1/48 (2%)
Query 49 ASAVLVLDGRGKVIIGRDYRGDVSLAISERFACRVID-QEEAAVTPVF 95
ASAV VLD +GKV+I R+YRGDV ++ E F +++ +EE ++P+
Sbjct 3 ASAVYVLDLKGKVLICRNYRGDVDMSEVEHFMPILMEKEEEGMLSPIL 50
> Hs9506797
Length=423
Score = 43.1 bits (100), Expect = 1e-04, Method: Composition-based stats.
Identities = 20/48 (41%), Positives = 33/48 (68%), Gaps = 1/48 (2%)
Query 49 ASAVLVLDGRGKVIIGRDYRGDVSLAISERFACRVID-QEEAAVTPVF 95
ASAV +LD +GK +I R+Y+GDV+++ E F ++ +EE A+ P+
Sbjct 3 ASAVFILDVKGKPLISRNYKGDVAMSKIEHFMPLLVQREEEGALAPLL 50
> CE11129
Length=470
Score = 42.7 bits (99), Expect = 2e-04, Method: Composition-based stats.
Identities = 18/47 (38%), Positives = 32/47 (68%), Gaps = 1/47 (2%)
Query 50 SAVLVLDGRGKVIIGRDYRGDVSLAISERFACRVIDQE-EAAVTPVF 95
S + +LD +G V+I R+YRGDV ++ E+F ++++E E + +PV
Sbjct 4 SGLFILDLKGNVVISRNYRGDVDMSCIEKFMPLLVEKEDEGSASPVL 50
> CE28050
Length=422
Score = 41.6 bits (96), Expect = 4e-04, Method: Composition-based stats.
Identities = 18/49 (36%), Positives = 31/49 (63%), Gaps = 1/49 (2%)
Query 48 GASAVLVLDGRGKVIIGRDYRGDVSLAISERFACRVID-QEEAAVTPVF 95
SA+ +LD +GK II R+YRGD+ + ++F +++ +EE + PV
Sbjct 2 ATSAMFILDLKGKTIISRNYRGDIDMTAIDKFIHLLMEKEEEGSAAPVL 50
> At5g46630
Length=438
Score = 33.9 bits (76), Expect = 0.069, Method: Composition-based stats.
Identities = 17/47 (36%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 48 GASAVLVLDGRGKVIIGRDYRGDVSLAISERFACRVIDQEEAAVTPV 94
ASA+ L+ RG V+I R YR DV + + F ++ +E PV
Sbjct 4 AASAIYFLNLRGDVLINRTYRDDVGGNMVDAFRTHIMQTKELGNCPV 50
> YPL259c
Length=475
Score = 32.7 bits (73), Expect = 0.15, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 49 ASAVLVLDGRGKVIIGRDYRGDVSLAISERFACRVIDQEEAA 90
ASAV D GK ++ R YR D+ L+ ++F + D EE +
Sbjct 2 ASAVYFCDHNGKPLLSRRYRDDIPLSAIDKFPILLSDLEEQS 43
> CE28642
Length=482
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 14/44 (31%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 51 AVLVLDGRGKVIIGRDYRGDVSLAISERFACRVIDQEEAAVTPV 94
+ V + +G+V+I R YR DV+ + F VI + +PV
Sbjct 4 GLFVYNHKGEVLISRIYRDDVTRNAVDAFRVNVIHARQQVRSPV 47
> YJR130c
Length=639
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 14/32 (43%), Positives = 23/32 (71%), Gaps = 3/32 (9%)
Query 30 VLCLSLRRLFAGCSAKMAGASAVLVLDGRGKV 61
++C SL ++F+G S +AG+ LVL+ RGK+
Sbjct 436 IVCSSLTKIFSGDSNVIAGS---LVLNPRGKI 464
> Hs4557775
Length=166
Score = 28.5 bits (62), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 50 SAVLVLDGRGKVIIGRDYRGDVSLAISERFACRVIDQEEAAVTP 93
+A V D GK ++ DY ++ +ERF+ +DQ AA P
Sbjct 101 NAFKVFDPEGKGVLKADYVREMLTTQAERFSKEEVDQMFAAFPP 144
> Hs17475023
Length=166
Score = 28.5 bits (62), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 50 SAVLVLDGRGKVIIGRDYRGDVSLAISERFACRVIDQEEAAVTP 93
+A V D GK ++ DY ++ +ERF+ +DQ AA P
Sbjct 101 NAFKVFDPEGKGVLKADYVREMLTTQAERFSKEEVDQMFAAFPP 144
> 7300861
Length=438
Score = 28.1 bits (61), Expect = 4.7, Method: Composition-based stats.
Identities = 13/44 (29%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 51 AVLVLDGRGKVIIGRDYRGDVSLAISERFACRVIDQEEAAVTPV 94
+ V + +G+V+I R YR D+ + F VI + +PV
Sbjct 4 GLFVYNHKGEVLISRVYRDDIGRNAVDAFRVNVIHARQQVRSPV 47
> Hs4506075
Length=912
Score = 27.7 bits (60), Expect = 5.0, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 1/51 (1%)
Query 38 LFAGCSAKMAGASAVLVLD-GRGKVIIGRDYRGDVSLAISERFACRVIDQE 87
A +A + G S L + R V++ +D GD SLA AC ++DQ+
Sbjct 38 FLAPVAAPVGGISFHLQIGLSREPVLLLQDSSGDYSLAHVREMACSIVDQK 88
> YOL062c
Length=491
Score = 26.9 bits (58), Expect = 9.7, Method: Composition-based stats.
Identities = 11/36 (30%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 50 SAVLVLDGRGKVIIGRDYRGDVSLAISERFACRVID 85
S VLV RG++++ + ++ + +IS+ F +VI+
Sbjct 3 SGVLVYSSRGELVLNKFFKNSLKRSISDIFRVQVIN 38
Lambda K H
0.325 0.133 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194657780
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40