bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1632_orf1
Length=155
Score E
Sequences producing significant alignments: (Bits) Value
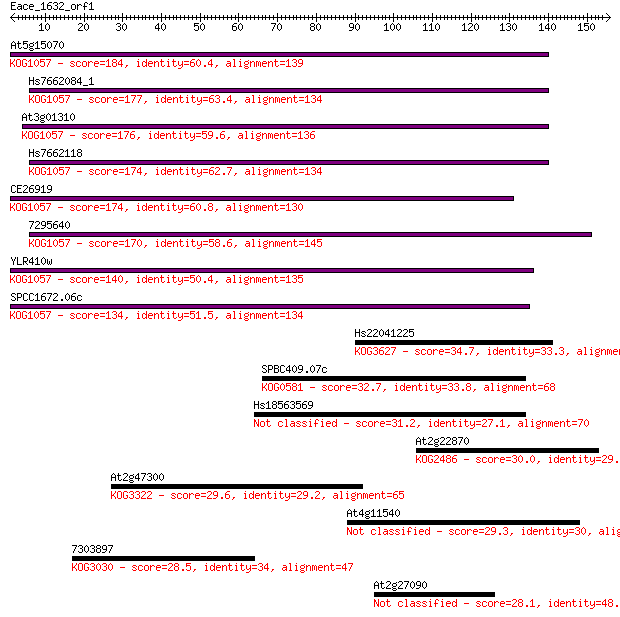
At5g15070 184 7e-47
Hs7662084_1 177 9e-45
At3g01310 176 1e-44
Hs7662118 174 5e-44
CE26919 174 7e-44
7295640 170 1e-42
YLR410w 140 1e-33
SPCC1672.06c 134 1e-31
Hs22041225 34.7 0.077
SPBC409.07c 32.7 0.30
Hs18563569 31.2 0.85
At2g22870 30.0 1.9
At2g47300 29.6 2.7
At4g11540 29.3 3.6
7303897 28.5 5.1
At2g27090 28.1 6.8
> At5g15070
Length=1030
Score = 184 bits (467), Expect = 7e-47, Method: Composition-based stats.
Identities = 84/139 (60%), Positives = 109/139 (78%), Gaps = 0/139 (0%)
Query 1 KQQNSSSSYFPDIHQVRKDGVYLYEEFLPTFGTDVKVYTVGPLFAHAEARKSPSVDGVVC 60
K N SS + PD+ +VR++G Y+YEEF+PT GTDVKVYTVGP +AHAEARKSP VDGVV
Sbjct 170 KVGNRSSEFHPDVRRVRREGSYIYEEFMPTGGTDVKVYTVGPEYAHAEARKSPVVDGVVM 229
Query 61 RSADGKEIRYPVILTEFEKCIAFKLVRAFKQVVCGFDILRTSRGPFVCDVNGFSFVKSSI 120
R+ DGKE+RYPV+LT EK +A ++ AF+Q VCGFD+LR+ +VCDVNG+SFVK+S
Sbjct 230 RNPDGKEVRYPVLLTPAEKQMAREVCIAFRQAVCGFDLLRSEGSSYVCDVNGWSFVKNSY 289
Query 121 KYYEDCSSIIRLYFLNKLA 139
KYY+D + ++R FL+ A
Sbjct 290 KYYDDAACVLRKMFLDAKA 308
> Hs7662084_1
Length=1099
Score = 177 bits (448), Expect = 9e-45, Method: Composition-based stats.
Identities = 85/134 (63%), Positives = 105/134 (78%), Gaps = 1/134 (0%)
Query 6 SSSYFPDIHQVRKDGVYLYEEFLPTFGTDVKVYTVGPLFAHAEARKSPSVDGVVCRSADG 65
SS Y P+ VRK G Y+YEEF+PT GTDVKVYTVGP +AHAEARKSP++DG V R ++G
Sbjct 230 SSVYSPE-SSVRKTGSYIYEEFMPTDGTDVKVYTVGPDYAHAEARKSPALDGKVERDSEG 288
Query 66 KEIRYPVILTEFEKCIAFKLVRAFKQVVCGFDILRTSRGPFVCDVNGFSFVKSSIKYYED 125
KEIRYPV+LT EK +A K+ AFKQ VCGFD+LR + FVCDVNGFSFVK+S+KYY+D
Sbjct 289 KEIRYPVMLTAMEKLVARKVCVAFKQTVCGFDLLRANGHSFVCDVNGFSFVKNSMKYYDD 348
Query 126 CSSIIRLYFLNKLA 139
C+ I+ + +LA
Sbjct 349 CAKILGNTIMRELA 362
> At3g01310
Length=1015
Score = 176 bits (447), Expect = 1e-44, Method: Composition-based stats.
Identities = 81/136 (59%), Positives = 106/136 (77%), Gaps = 0/136 (0%)
Query 4 NSSSSYFPDIHQVRKDGVYLYEEFLPTFGTDVKVYTVGPLFAHAEARKSPSVDGVVCRSA 63
N SS + PD+ +VR++G Y+YEEF+ T GTDVKVYTVGP +AHAEARKSP VDGVV R+
Sbjct 156 NRSSEFHPDVRRVRREGSYIYEEFMATGGTDVKVYTVGPEYAHAEARKSPVVDGVVMRNT 215
Query 64 DGKEIRYPVILTEFEKCIAFKLVRAFKQVVCGFDILRTSRGPFVCDVNGFSFVKSSIKYY 123
DGKE+RYPV+LT EK +A ++ AF+Q VCGFD+LR+ +VCDVNG+SFVK+S KYY
Sbjct 216 DGKEVRYPVLLTPAEKQMAREVCIAFRQAVCGFDLLRSEGCSYVCDVNGWSFVKNSYKYY 275
Query 124 EDCSSIIRLYFLNKLA 139
+D + ++R L+ A
Sbjct 276 DDAACVLRKMCLDAKA 291
> Hs7662118
Length=1243
Score = 174 bits (442), Expect = 5e-44, Method: Compositional matrix adjust.
Identities = 84/134 (62%), Positives = 105/134 (78%), Gaps = 1/134 (0%)
Query 6 SSSYFPDIHQVRKDGVYLYEEFLPTFGTDVKVYTVGPLFAHAEARKSPSVDGVVCRSADG 65
SS Y P+ VRK G Y+YEEF+PT GTDVKVYTVGP +AHAEARKSP++DG V R ++G
Sbjct 219 SSVYSPE-SNVRKTGSYIYEEFMPTDGTDVKVYTVGPDYAHAEARKSPALDGKVERDSEG 277
Query 66 KEIRYPVILTEFEKCIAFKLVRAFKQVVCGFDILRTSRGPFVCDVNGFSFVKSSIKYYED 125
KE+RYPVIL EK IA+K+ AFKQ VCGFD+LR + +VCDVNGFSFVK+S+KYY+D
Sbjct 278 KEVRYPVILNAREKLIAWKVCLAFKQTVCGFDLLRANGQSYVCDVNGFSFVKNSMKYYDD 337
Query 126 CSSIIRLYFLNKLA 139
C+ I+ + +LA
Sbjct 338 CAKILGNIVMRELA 351
> CE26919
Length=1339
Score = 174 bits (440), Expect = 7e-44, Method: Composition-based stats.
Identities = 79/130 (60%), Positives = 101/130 (77%), Gaps = 0/130 (0%)
Query 1 KQQNSSSSYFPDIHQVRKDGVYLYEEFLPTFGTDVKVYTVGPLFAHAEARKSPSVDGVVC 60
++ N+ SS++ +VRK+G Y+YEEF+P GTDVKVY VGP +AHAEARK+P +DG V
Sbjct 188 RKINNRSSWYSPKSEVRKEGSYIYEEFIPADGTDVKVYAVGPFYAHAEARKAPGLDGKVE 247
Query 61 RSADGKEIRYPVILTEFEKCIAFKLVRAFKQVVCGFDILRTSRGPFVCDVNGFSFVKSSI 120
R +DGKE+RYPVIL+ EK IA K+V AF Q VCGFD+LR + +VCDVNGFSFVK+S
Sbjct 248 RDSDGKEVRYPVILSNKEKQIAKKIVLAFGQTVCGFDLLRANGKSYVCDVNGFSFVKTST 307
Query 121 KYYEDCSSII 130
KYYED + I+
Sbjct 308 KYYEDTAKIL 317
> 7295640
Length=1751
Score = 170 bits (430), Expect = 1e-42, Method: Composition-based stats.
Identities = 85/145 (58%), Positives = 106/145 (73%), Gaps = 6/145 (4%)
Query 6 SSSYFPDIHQVRKDGVYLYEEFLPTFGTDVKVYTVGPLFAHAEARKSPSVDGVVCRSADG 65
SS Y P+ +VRK G ++YE+F+PT GTDVKVYTVGP +AHAEARKSP++DG V R ++G
Sbjct 242 SSVYSPE-SRVRKTGSFIYEDFMPTDGTDVKVYTVGPDYAHAEARKSPALDGKVERDSEG 300
Query 66 KEIRYPVILTEFEKCIAFKLVRAFKQVVCGFDILRTSRGPFVCDVNGFSFVKSSIKYYED 125
KEIRYPVIL EK I+ K+ AFKQ VCGFD+LR + +VCDVNGFSFVK+S KYY+D
Sbjct 301 KEIRYPVILNHSEKLISRKVCLAFKQTVCGFDLLRANGKSYVCDVNGFSFVKNSNKYYDD 360
Query 126 CSSIIRLYFLNKLAYRCCRAALHAP 150
C+ I+ L +L LH P
Sbjct 361 CAKILGNMILRELT-----PTLHIP 380
> YLR410w
Length=1146
Score = 140 bits (352), Expect = 1e-33, Method: Composition-based stats.
Identities = 68/136 (50%), Positives = 92/136 (67%), Gaps = 1/136 (0%)
Query 1 KQQNSSSSYFPDIHQVRKDGVYLYEEFLPTFG-TDVKVYTVGPLFAHAEARKSPSVDGVV 59
K N SS + P + R +G Y+YE+F+ T DVK YT+G F HAE RKSP VDG+V
Sbjct 378 KVGNKSSEFDPTLVHPRTEGSYIYEQFMDTDNFEDVKAYTIGENFCHAETRKSPVVDGIV 437
Query 60 CRSADGKEIRYPVILTEFEKCIAFKLVRAFKQVVCGFDILRTSRGPFVCDVNGFSFVKSS 119
R+ GKE+RY L++ EK IA K+ +AF Q++CGFD+LR S +V DVNGFSFVK +
Sbjct 438 RRNTHGKEVRYITELSDEEKTIAGKVSKAFSQMICGFDLLRVSGKSYVIDVNGFSFVKDN 497
Query 120 IKYYEDCSSIIRLYFL 135
YY+ C++I+R F+
Sbjct 498 KAYYDSCANILRSTFI 513
> SPCC1672.06c
Length=920
Score = 134 bits (336), Expect = 1e-31, Method: Composition-based stats.
Identities = 69/135 (51%), Positives = 92/135 (68%), Gaps = 1/135 (0%)
Query 1 KQQNSSSSYFPDIHQVRKDGVYLYEEFLPT-FGTDVKVYTVGPLFAHAEARKSPSVDGVV 59
K N SS Y PD+ R +G ++YEEF+ DVKVYTVGP ++HAE RKSP VDG+V
Sbjct 224 KVANKSSDYDPDLCAPRTEGSFIYEEFMNVDNAEDVKVYTVGPHYSHAETRKSPVVDGIV 283
Query 60 CRSADGKEIRYPVILTEFEKCIAFKLVRAFKQVVCGFDILRTSRGPFVCDVNGFSFVKSS 119
R+ GKEIR+ L+E EK +A K+ AF+Q VCGFD+LR S +V DVNG+SFVK +
Sbjct 284 RRNPHGKEIRFITNLSEEEKNMASKISIAFEQPVCGFDLLRVSGQSYVIDVNGWSFVKDN 343
Query 120 IKYYEDCSSIIRLYF 134
YY++ + I++ F
Sbjct 344 NDYYDNAARILKQMF 358
> Hs22041225
Length=352
Score = 34.7 bits (78), Expect = 0.077, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 27/58 (46%), Gaps = 7/58 (12%)
Query 90 KQVVCGFDIL-------RTSRGPFVCDVNGFSFVKSSIKYYEDCSSIIRLYFLNKLAY 140
+ ++C D++ R SRGP VC +NG F+ + DC S + +L Y
Sbjct 232 EDMLCAGDLITGKAICRRDSRGPLVCPLNGTWFLMGLSSWSLDCCSPVGPRVFTRLPY 289
> SPBC409.07c
Length=605
Score = 32.7 bits (73), Expect = 0.30, Method: Composition-based stats.
Identities = 23/72 (31%), Positives = 34/72 (47%), Gaps = 4/72 (5%)
Query 66 KEIRYPVILTEFEKCIAFKLVRA-FKQVVCGFDILRTSRGPFVCDVNGFSFVKSSI---K 121
K + P +T K I L A F Q++ DIL + P++ D G FV+ S+
Sbjct 336 KALHQPTGVTMALKEIRLSLEEATFNQIIMELDILHKAVSPYIVDFYGAFFVEGSVFICM 395
Query 122 YYEDCSSIIRLY 133
Y D S+ +LY
Sbjct 396 EYMDAGSMDKLY 407
> Hs18563569
Length=217
Score = 31.2 bits (69), Expect = 0.85, Method: Compositional matrix adjust.
Identities = 19/76 (25%), Positives = 35/76 (46%), Gaps = 6/76 (7%)
Query 64 DGKEIRYPVILTEFEKCIAFKLVRAFK--QVVCGFDILRTSRGPFVCDVNGFSFVKSSIK 121
D IR P+++ ++ + + Q +C FD+ R +R F+ N F ++ +K
Sbjct 111 DYPSIRPPILILSVKRWPGVSEQQVYHKFQNLCKFDVRRLTRSQFLLLTNKFKDARNILK 170
Query 122 YYED----CSSIIRLY 133
Y D C S+ R +
Sbjct 171 EYRDHPTLCISLYRYW 186
> At2g22870
Length=219
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 24/47 (51%), Gaps = 0/47 (0%)
Query 106 FVCDVNGFSFVKSSIKYYEDCSSIIRLYFLNKLAYRCCRAALHAPLP 152
++ D+ G+ F K S D S+ + YFLN+ + C + A +P
Sbjct 88 YIVDLPGYGFAKVSDAAKTDWSAFTKGYFLNRDSLVCVLLLIDASVP 134
> At2g47300
Length=344
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 33/65 (50%), Gaps = 7/65 (10%)
Query 27 FLPTFGTDVKVYTVGPLFAHAEARKSPSVDGVVCRSADGKEIRYPVILTEFEKCIAFKLV 86
FL TF +D + LF H ++ PS D ++ D K++R + + + C+ L+
Sbjct 264 FLQTFTSDNML-----LFPHNTSK--PSTDLMMTLQEDDKKVRAQIHQSSNKLCLVRVLL 316
Query 87 RAFKQ 91
AFK+
Sbjct 317 HAFKE 321
> At4g11540
Length=525
Score = 29.3 bits (64), Expect = 3.6, Method: Composition-based stats.
Identities = 18/60 (30%), Positives = 30/60 (50%), Gaps = 10/60 (16%)
Query 88 AFKQVVCGFDILRTSRGPFVCDVNGFSFVKSSIKYYEDCSSIIRLYFLNKLAYRCCRAAL 147
+F CG L+ R P++C GF ++DC S+ RL +N+ A+R R ++
Sbjct 167 SFTCNACG---LKGDRSPYICVQCGFII-------HQDCLSLPRLININRHAHRVSRTSV 216
> 7303897
Length=372
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 23/48 (47%), Gaps = 1/48 (2%)
Query 17 RKDGVYLYEEFLPTFGTDVKVYTVGPLFAHAEARKSPSV-DGVVCRSA 63
+K G+Y + L TD+ Y++G L H A P + DG C A
Sbjct 184 KKIGIYAFGAVLSQLTTDIAKYSIGRLRPHFIAVCQPQMADGSTCDDA 231
> At2g27090
Length=743
Score = 28.1 bits (61), Expect = 6.8, Method: Composition-based stats.
Identities = 15/31 (48%), Positives = 19/31 (61%), Gaps = 2/31 (6%)
Query 95 GFDILRTSRGPFVCDVNGFSFVKSSIKYYED 125
GFD RTS FV +N F +SS+K YE+
Sbjct 691 GFDRFRTSLEGFVGQLN--QFAESSVKMYEE 719
Lambda K H
0.325 0.140 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2033998736
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40