bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1583_orf1
Length=165
Score E
Sequences producing significant alignments: (Bits) Value
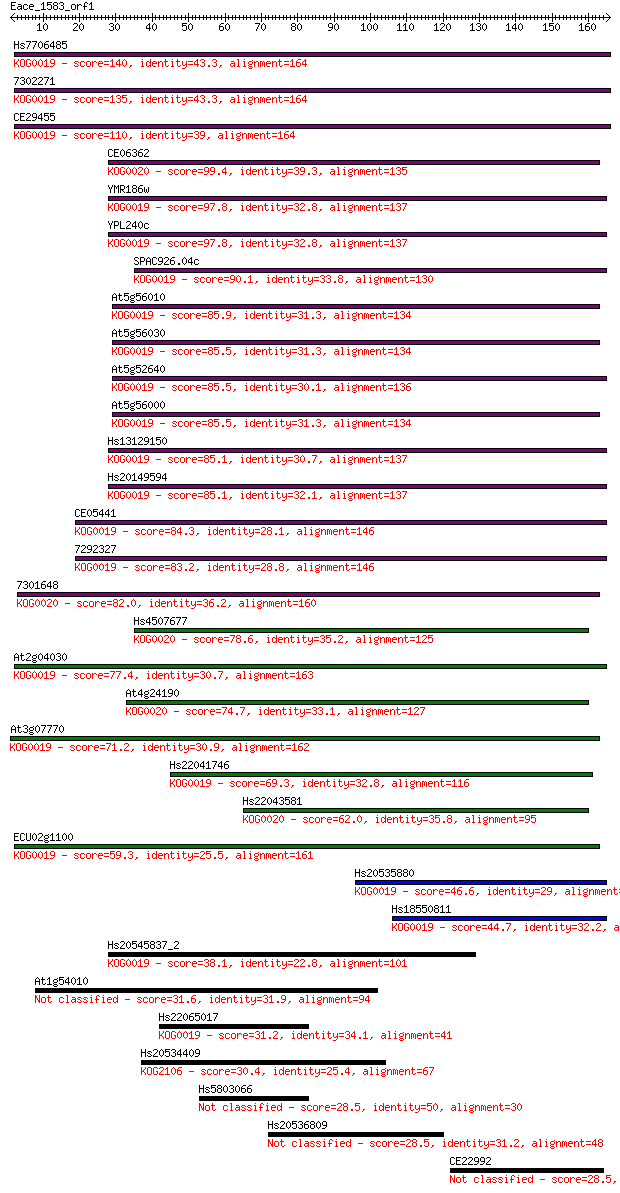
Hs7706485 140 1e-33
7302271 135 4e-32
CE29455 110 1e-24
CE06362 99.4 3e-21
YMR186w 97.8 8e-21
YPL240c 97.8 9e-21
SPAC926.04c 90.1 2e-18
At5g56010 85.9 4e-17
At5g56030 85.5 4e-17
At5g52640 85.5 5e-17
At5g56000 85.5 5e-17
Hs13129150 85.1 5e-17
Hs20149594 85.1 6e-17
CE05441 84.3 9e-17
7292327 83.2 2e-16
7301648 82.0 5e-16
Hs4507677 78.6 5e-15
At2g04030 77.4 1e-14
At4g24190 74.7 7e-14
At3g07770 71.2 8e-13
Hs22041746 69.3 3e-12
Hs22043581 62.0 5e-10
ECU02g1100 59.3 3e-09
Hs20535880 46.6 2e-05
Hs18550811 44.7 8e-05
Hs20545837_2 38.1 0.007
At1g54010 31.6 0.81
Hs22065017 31.2 0.89
Hs20534409 30.4 1.6
Hs5803066 28.5 6.6
Hs20536809 28.5 6.7
CE22992 28.5 6.8
> Hs7706485
Length=704
Score = 140 bits (352), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 71/166 (42%), Positives = 114/166 (68%), Gaps = 7/166 (4%)
Query 2 RVKECAQKFSAFVNFPVYMEDNGKEVQVSSQEALWLKTSATPEE--HKQFFRHLTNQSWG 59
RV++ K+S FV+FP+Y+ NG+ ++++ +A+W+ E H++F+R++ Q+
Sbjct 269 RVRDVVTKYSNFVSFPLYL--NGR--RMNTLQAIWMMDPKDVGEWQHEEFYRYVA-QAHD 323
Query 60 EPFYSIMFSADAPLSIRSVLYFPADPPNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKW 119
+P Y++ + DAPL+IRS+ Y P P+ + L S V+L SR VL++ ATD+LPKW
Sbjct 324 KPRYTLHYKTDAPLNIRSIFYVPDMKPSMFDVSRELGSSVALYSRKVLIQTKATDILPKW 383
Query 120 LWFLRGIVDCEDLPLNVSREHMQDSALQRKLSSVIVRRILRFLNDQ 165
L F+RG+VD ED+PLN+SRE +Q+SAL RKL V+ +R+++F DQ
Sbjct 384 LRFIRGVVDSEDIPLNLSRELLQESALIRKLRDVLQQRLIKFFIDQ 429
> 7302271
Length=691
Score = 135 bits (340), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 71/166 (42%), Positives = 109/166 (65%), Gaps = 7/166 (4%)
Query 2 RVKECAQKFSAFVNFPVYMEDNGKEVQVSSQEALWL--KTSATPEEHKQFFRHLTNQSWG 59
R+K +K+S FV P+ + NGK Q + + LWL S + E+H F+R ++N S+
Sbjct 257 RIKAVIKKYSNFVGSPILL--NGK--QANEIKPLWLLEPQSISKEQHHDFYRFISN-SFD 311
Query 60 EPFYSIMFSADAPLSIRSVLYFPADPPNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKW 119
P +++ ++AD PLSI ++LYFP P + +GV+L +R VL++ LLPKW
Sbjct 312 VPRFTLHYNADVPLSIHALLYFPEGKPGLFEMSRDGNTGVALYTRKVLIQSKTEHLLPKW 371
Query 120 LWFLRGIVDCEDLPLNVSREHMQDSALQRKLSSVIVRRILRFLNDQ 165
L F++G+VD ED+PLN+SRE +Q+S+L RKLSSVI R++RFL ++
Sbjct 372 LRFVKGVVDSEDIPLNLSRELLQNSSLIRKLSSVISTRVIRFLQER 417
> CE29455
Length=657
Score = 110 bits (274), Expect = 1e-24, Method: Composition-based stats.
Identities = 64/172 (37%), Positives = 101/172 (58%), Gaps = 12/172 (6%)
Query 2 RVKECAQKFSAFVNFPVYMEDNGKEVQVSSQEALWLKTS--ATPEEHKQFFRHLT----- 54
R+KE K+S FV+ P+ + NG+ +V++ A+W + E H+ FF+ L
Sbjct 210 RIKEVINKYSYFVSAPILV--NGE--RVNNLNAIWTMQAREVNKEMHETFFKQLVKTQGK 265
Query 55 NQSWGEPFYSIMFSADAPLSIRSVLYFPADPPNRL-FQTGPLESGVSLLSRPVLVKKSAT 113
+ + P Y+I F D P+S+RSV+Y P N+L F G+SL +R VL+K A
Sbjct 266 QEMYTRPQYTIHFQTDTPVSLRSVIYIPQTQFNQLTFMAQQTMCGLSLYARRVLIKPDAQ 325
Query 114 DLLPKWLWFLRGIVDCEDLPLNVSREHMQDSALQRKLSSVIVRRILRFLNDQ 165
+L+P +L F+ G+VD ED+PLN+SRE +Q++ + RKL +I +IL L +
Sbjct 326 ELIPNYLRFVIGVVDSEDIPLNLSREMLQNNPVLRKLRKIITDKILGSLQSE 377
> CE06362
Length=760
Score = 99.4 bits (246), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 53/138 (38%), Positives = 84/138 (60%), Gaps = 4/138 (2%)
Query 28 QVSSQEALWLK--TSATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADP 85
+V++ + +W++ +E+KQF++ +T S EP + FSA+ +S RS+LY P
Sbjct 314 KVNNVKPIWMRKPNQVEEDEYKQFYKSITKDS-EEPLSHVHFSAEGEVSFRSILYVPKKS 372
Query 86 PNRLFQT-GPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDS 144
PN +FQ G + + L R V + D+LPK+L F+RGIVD +DLPLNVSRE++Q
Sbjct 373 PNDMFQNYGKVIENIKLYVRRVFITDDFADMLPKYLSFIRGIVDSDDLPLNVSRENLQQH 432
Query 145 ALQRKLSSVIVRRILRFL 162
L + + +VR++L L
Sbjct 433 KLLKVIKKKLVRKVLDML 450
> YMR186w
Length=705
Score = 97.8 bits (242), Expect = 8e-21, Method: Composition-based stats.
Identities = 45/139 (32%), Positives = 83/139 (59%), Gaps = 3/139 (2%)
Query 28 QVSSQEALWLKTSA--TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADP 85
+++ + LW + + T EE+ F++ ++N W +P Y FS + L R++L+ P
Sbjct 265 ELNKTKPLWTRNPSDITQEEYNAFYKSISN-DWEDPLYVKHFSVEGQLEFRAILFIPKRA 323
Query 86 PNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSA 145
P LF++ ++ + L R V + A DL+P+WL F++G+VD EDLPLN+SRE +Q +
Sbjct 324 PFDLFESKKKKNNIKLYVRRVFITDEAEDLIPEWLSFVKGVVDSEDLPLNLSREMLQQNK 383
Query 146 LQRKLSSVIVRRILRFLND 164
+ + + IV++++ N+
Sbjct 384 IMKVIRKNIVKKLIEAFNE 402
> YPL240c
Length=709
Score = 97.8 bits (242), Expect = 9e-21, Method: Composition-based stats.
Identities = 45/139 (32%), Positives = 83/139 (59%), Gaps = 3/139 (2%)
Query 28 QVSSQEALWLKTSA--TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADP 85
+++ + LW + + T EE+ F++ ++N W +P Y FS + L R++L+ P
Sbjct 269 ELNKTKPLWTRNPSDITQEEYNAFYKSISN-DWEDPLYVKHFSVEGQLEFRAILFIPKRA 327
Query 86 PNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSA 145
P LF++ ++ + L R V + A DL+P+WL F++G+VD EDLPLN+SRE +Q +
Sbjct 328 PFDLFESKKKKNNIKLYVRRVFITDEAEDLIPEWLSFVKGVVDSEDLPLNLSREMLQQNK 387
Query 146 LQRKLSSVIVRRILRFLND 164
+ + + IV++++ N+
Sbjct 388 IMKVIRKNIVKKLIEAFNE 406
> SPAC926.04c
Length=704
Score = 90.1 bits (222), Expect = 2e-18, Method: Composition-based stats.
Identities = 44/132 (33%), Positives = 74/132 (56%), Gaps = 3/132 (2%)
Query 35 LWLK--TSATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPPNRLFQT 92
+W + + T EE+ F++ LTN W + FS + L R++L+ P P LF+
Sbjct 271 IWTRNPSEVTKEEYASFYKSLTN-DWEDHLAVKHFSVEGQLEFRAILFVPRRAPMDLFEA 329
Query 93 GPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSALQRKLSS 152
++ + L R V + +L+P+WL F++G+VD EDLPLN+SRE +Q + + + +
Sbjct 330 KRKKNNIKLYVRRVFITDDCEELIPEWLGFIKGVVDSEDLPLNLSREMLQQNKIMKVIRK 389
Query 153 VIVRRILRFLND 164
+VRR L N+
Sbjct 390 NLVRRCLDMFNE 401
> At5g56010
Length=699
Score = 85.9 bits (211), Expect = 4e-17, Method: Composition-based stats.
Identities = 42/136 (30%), Positives = 75/136 (55%), Gaps = 3/136 (2%)
Query 29 VSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPP 86
V+ Q+ +W++ EE+ F++ L+N W E FS + L +++L+ P P
Sbjct 261 VNKQKPIWMRKPEEINKEEYAAFYKSLSN-DWEEHLAVKHFSVEGQLEFKAILFVPKRAP 319
Query 87 NRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSAL 146
LF T + + L R V + + D++P++L F++GIVD EDLPLN+SRE +Q + +
Sbjct 320 FDLFDTKKKPNNIKLYVRRVFIMDNCEDIIPEYLGFVKGIVDSEDLPLNISRETLQQNKI 379
Query 147 QRKLSSVIVRRILRFL 162
+ + +V++ L
Sbjct 380 LKVIRKNLVKKCLELF 395
> At5g56030
Length=699
Score = 85.5 bits (210), Expect = 4e-17, Method: Composition-based stats.
Identities = 42/136 (30%), Positives = 75/136 (55%), Gaps = 3/136 (2%)
Query 29 VSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPP 86
V+ Q+ +W++ EE+ F++ L+N W E FS + L +++L+ P P
Sbjct 261 VNKQKPIWMRKPEEINKEEYAAFYKSLSN-DWEEHLAVKHFSVEGQLEFKAILFVPKRAP 319
Query 87 NRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSAL 146
LF T + + L R V + + D++P++L F++GIVD EDLPLN+SRE +Q + +
Sbjct 320 FDLFDTKKKPNNIKLYVRRVFIMDNCEDIIPEYLGFVKGIVDSEDLPLNISRETLQQNKI 379
Query 147 QRKLSSVIVRRILRFL 162
+ + +V++ L
Sbjct 380 LKVIRKNLVKKCLELF 395
> At5g52640
Length=705
Score = 85.5 bits (210), Expect = 5e-17, Method: Composition-based stats.
Identities = 41/138 (29%), Positives = 78/138 (56%), Gaps = 3/138 (2%)
Query 29 VSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPP 86
++ Q+ +WL+ T EE+ F++ LTN W + FS + L +++L+ P P
Sbjct 267 INKQKPIWLRKPEEITKEEYAAFYKSLTN-DWEDHLAVKHFSVEGQLEFKAILFVPKRAP 325
Query 87 NRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSAL 146
LF T + + L R V + + +L+P++L F++G+VD +DLPLN+SRE +Q + +
Sbjct 326 FDLFDTRKKLNNIKLYVRRVFIMDNCEELIPEYLSFVKGVVDSDDLPLNISRETLQQNKI 385
Query 147 QRKLSSVIVRRILRFLND 164
+ + +V++ + N+
Sbjct 386 LKVIRKNLVKKCIEMFNE 403
> At5g56000
Length=699
Score = 85.5 bits (210), Expect = 5e-17, Method: Composition-based stats.
Identities = 42/136 (30%), Positives = 74/136 (54%), Gaps = 3/136 (2%)
Query 29 VSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPP 86
V+ Q+ +W++ EE+ F++ L+N W E FS + L +++L+ P P
Sbjct 261 VNKQKPIWMRKPEEINKEEYAAFYKSLSN-DWEEHLAVKHFSVEGQLEFKAILFVPKRAP 319
Query 87 NRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSAL 146
LF T + + L R V + + D++P +L F++GIVD EDLPLN+SRE +Q + +
Sbjct 320 FDLFDTKKKPNNIKLYVRRVFIMDNCEDIIPDYLGFVKGIVDSEDLPLNISRETLQQNKI 379
Query 147 QRKLSSVIVRRILRFL 162
+ + +V++ L
Sbjct 380 LKVIRKNLVKKCLELF 395
> Hs13129150
Length=732
Score = 85.1 bits (209), Expect = 5e-17, Method: Composition-based stats.
Identities = 42/139 (30%), Positives = 78/139 (56%), Gaps = 3/139 (2%)
Query 28 QVSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADP 85
+++ + +W + T EE+ +F++ LTN W + FS + L R++L+ P
Sbjct 289 ELNKTKPIWTRNPDDITNEEYGEFYKSLTN-DWEDHLAVKHFSVEGQLEFRALLFVPRRA 347
Query 86 PNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSA 145
P LF+ ++ + L R V + + +L+P++L F+RG+VD EDLPLN+SRE +Q S
Sbjct 348 PFDLFENRKKKNNIKLYVRRVFIMDNCEELIPEYLNFIRGVVDSEDLPLNISREMLQQSK 407
Query 146 LQRKLSSVIVRRILRFLND 164
+ + + +V++ L +
Sbjct 408 ILKVIRKNLVKKCLELFTE 426
> Hs20149594
Length=724
Score = 85.1 bits (209), Expect = 6e-17, Method: Composition-based stats.
Identities = 44/139 (31%), Positives = 79/139 (56%), Gaps = 3/139 (2%)
Query 28 QVSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADP 85
+++ + +W + T EE+ +F++ LTN W + FS + L R++L+ P
Sbjct 281 ELNKTKPIWTRNPDDITQEEYGEFYKSLTN-DWEDHLAVKHFSVEGQLEFRALLFIPRRA 339
Query 86 PNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSA 145
P LF+ ++ + L R V + S +L+P++L F+RG+VD EDLPLN+SRE +Q S
Sbjct 340 PFDLFENKKKKNNIKLYVRRVFIMDSCDELIPEYLNFIRGVVDSEDLPLNISREMLQQSK 399
Query 146 LQRKLSSVIVRRILRFLND 164
+ + + IV++ L ++
Sbjct 400 ILKVIRKNIVKKCLELFSE 418
> CE05441
Length=702
Score = 84.3 bits (207), Expect = 9e-17, Method: Composition-based stats.
Identities = 41/148 (27%), Positives = 84/148 (56%), Gaps = 7/148 (4%)
Query 19 YMEDNGKEVQVSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIR 76
Y ED +++ + +W + + EE+ +F++ L+N W + FS + L R
Sbjct 255 YFEDE----ELNKTKPIWTRNPDDISNEEYAEFYKSLSN-DWEDHLAVKHFSVEGQLEFR 309
Query 77 SVLYFPADPPNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNV 136
++L+ P P LF+ ++ + L R V + ++ +L+P++L F++G+VD EDLPLN+
Sbjct 310 ALLFVPQRAPFDLFENKKSKNSIKLYVRRVFIMENCEELMPEYLNFIKGVVDSEDLPLNI 369
Query 137 SREHMQDSALQRKLSSVIVRRILRFLND 164
SRE +Q S + + + +V++ + +++
Sbjct 370 SREMLQQSKILKVIRKNLVKKCMELIDE 397
> 7292327
Length=717
Score = 83.2 bits (204), Expect = 2e-16, Method: Composition-based stats.
Identities = 42/148 (28%), Positives = 81/148 (54%), Gaps = 7/148 (4%)
Query 19 YMEDNGKEVQVSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIR 76
Y ED +++ + +W + + EE+ +F++ LTN W + FS + L R
Sbjct 269 YTEDE----ELNKTKPIWTRNPDDISQEEYGEFYKSLTN-DWEDHLAVKHFSVEGQLEFR 323
Query 77 SVLYFPADPPNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNV 136
++L+ P P LF+ + + L R V + + DL+P++L F++G+VD EDLPLN+
Sbjct 324 ALLFIPRRTPFDLFENQKKRNNIKLYVRRVFIMDNCEDLIPEYLNFMKGVVDSEDLPLNI 383
Query 137 SREHMQDSALQRKLSSVIVRRILRFLND 164
SRE +Q + + + + +V++ + + +
Sbjct 384 SREMLQQNKVLKVIRKNLVKKTMELIEE 411
> 7301648
Length=787
Score = 82.0 bits (201), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 58/209 (27%), Positives = 93/209 (44%), Gaps = 50/209 (23%)
Query 3 VKECAQKFSAFVNFPVYM-------------------------EDNGKEVQVSSQEA--- 34
V+E +K+S F+NFP+ M ED +V+ + E
Sbjct 264 VRELIRKYSQFINFPIRMWSSKTVEEEVPVEEEAKPEKSEDDVEDEDAKVEEAEDEKPKT 323
Query 35 ------------------LWLKTSA--TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLS 74
+W + A T +E+ F++ LT S EP F A+ ++
Sbjct 324 KKVSKTTWDWTLINDSKPIWTRKPAEVTEDEYTSFYKSLTKDS-SEPLTQTHFIAEGEVT 382
Query 75 IRSVLYFPADPPNRLF-QTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLP 133
+S+LY P P+ F + G + L R V + D++P +L F+RG+VD +DLP
Sbjct 383 FKSLLYVPKVQPSESFNRYGTKSDNIKLYVRRVFITDEFNDMMPNYLSFIRGVVDSDDLP 442
Query 134 LNVSREHMQDSALQRKLSSVIVRRILRFL 162
LNVSRE +Q L + + +VR++L L
Sbjct 443 LNVSRETLQQHKLIKVIKKKLVRKVLDML 471
> Hs4507677
Length=803
Score = 78.6 bits (192), Expect = 5e-15, Method: Composition-based stats.
Identities = 44/129 (34%), Positives = 75/129 (58%), Gaps = 5/129 (3%)
Query 35 LWLKTSATPEE--HKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPPNRLF-Q 91
+W + S EE +K F++ + +S +P I F+A+ ++ +S+L+ P P LF +
Sbjct 342 IWQRPSKEVEEDEYKAFYKSFSKES-DDPMAYIHFTAEGEVTFKSILFVPTSAPRGLFDE 400
Query 92 TGPLESG-VSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSALQRKL 150
G +S + L R V + D++PK+L F++G+VD +DLPLNVSRE +Q L + +
Sbjct 401 YGSKKSDYIKLYVRRVFITDDFHDMMPKYLNFVKGVVDSDDLPLNVSRETLQQHKLLKVI 460
Query 151 SSVIVRRIL 159
+VR+ L
Sbjct 461 RKKLVRKTL 469
> At2g04030
Length=780
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 50/199 (25%), Positives = 91/199 (45%), Gaps = 37/199 (18%)
Query 2 RVKECAQKFSAFVNFPVYM-EDNGKEVQVSSQEA-------------------------- 34
R+K + +S FV FP+Y ++ + ++V E
Sbjct 270 RIKNLVKNYSQFVGFPIYTWQEKSRTIEVEEDEPVKEGEEGEPKKKKTTKTEKYWDWELA 329
Query 35 -----LWLKTSATPE--EHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPA-DPP 86
LW++ S E E+ +F++ N+ + +P F+ + + RS+LY P P
Sbjct 330 NETKPLWMRNSKEVEKGEYNEFYKKAFNE-FLDPLAHTHFTTEGEVEFRSILYIPGMGPL 388
Query 87 NRLFQTGPLESGVSLLSRPVLVKKS-ATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSA 145
N T P + L + V + +L P++L F++G+VD +DLPLNVSRE +Q+S
Sbjct 389 NNEDVTNPKTKNIRLYVKRVFISDDFDGELFPRYLSFVKGVVDSDDLPLNVSREILQESR 448
Query 146 LQRKLSSVIVRRILRFLND 164
+ R + ++R+ + +
Sbjct 449 IVRIMRKRLIRKTFDMIQE 467
> At4g24190
Length=823
Score = 74.7 bits (182), Expect = 7e-14, Method: Composition-based stats.
Identities = 42/132 (31%), Positives = 75/132 (56%), Gaps = 5/132 (3%)
Query 33 EALWLKT--SATPEEHKQFFRHLTNQSWGE-PFYSIMFSADAPLSIRSVLYFPADPPNRL 89
+A+WL++ T EE+ +F+ L+ E P F+A+ + ++VLY P P+ L
Sbjct 345 KAIWLRSPKEVTEEEYTKFYHSLSKDFTDEKPMAWSHFNAEGDVEFKAVLYVPPKAPHDL 404
Query 90 FQT--GPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSALQ 147
+++ ++ + L R V + +LLPK+L FL+G+VD + LPLNVSRE +Q +
Sbjct 405 YESYYNSNKANLKLYVRRVFISDEFDELLPKYLSFLKGLVDSDTLPLNVSREMLQQHSSL 464
Query 148 RKLSSVIVRRIL 159
+ + ++R+ L
Sbjct 465 KTIKKKLIRKAL 476
> At3g07770
Length=803
Score = 71.2 bits (173), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 50/204 (24%), Positives = 90/204 (44%), Gaps = 43/204 (21%)
Query 1 HRVKECAQKFSAFVNFPVY------------MEDNGKEVQVSSQEA-------------- 34
R+++ + +S FV+FP+Y +ED+ E + Q+
Sbjct 286 ERIQKLVKNYSQFVSFPIYTWQEKGYTKEVEVEDDPTETKKDDQDDQTEKKKKTKKVVER 345
Query 35 ------------LWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLY 80
+WL+ T E+ +F+R N+ + +P S F+ + + RS+LY
Sbjct 346 YWDWELTNETQPIWLRNPKEVTTAEYNEFYRKAFNE-YLDPLASSHFTTEGEVEFRSILY 404
Query 81 FP-ADPPNRLFQTGPLESGVSLLSRPVLVKKS-ATDLLPKWLWFLRGIVDCEDLPLNVSR 138
P P + + L + V + +L P++L F++G+VD DLPLNVSR
Sbjct 405 VPPVSPSGKDDIVNQKTKNIRLYVKRVFISDDFDGELFPRYLSFVKGVVDSHDLPLNVSR 464
Query 139 EHMQDSALQRKLSSVIVRRILRFL 162
E +Q+S + R + +VR+ +
Sbjct 465 EILQESRIVRIMKKRLVRKAFDMI 488
> Hs22041746
Length=1595
Score = 69.3 bits (168), Expect = 3e-12, Method: Composition-based stats.
Identities = 38/116 (32%), Positives = 66/116 (56%), Gaps = 6/116 (5%)
Query 45 EHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPPNRLFQTGPLESGVSLLSR 104
E+++F++ LT +W + FS + L R+ L+ P P L +T ++ + L +R
Sbjct 762 EYREFYKSLT-INWEDYLAVKHFSVEGQLEFRAFLFVPRLAPFELLETRKKKNKIKLSAR 820
Query 105 PVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSALQRKLSSVIVRRILR 160
L+ + +L+P++L F+RG+VD EDLPLN+ RE A +S IV+R+ +
Sbjct 821 RDLIMDNCEELIPEYLNFIRGVVDSEDLPLNIFRETKDQVA-----NSTIVQRLWK 871
> Hs22043581
Length=329
Score = 62.0 bits (149), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 34/97 (35%), Positives = 57/97 (58%), Gaps = 6/97 (6%)
Query 65 IMFSADAPLSIRSVLYFPADPPNRLF-QTGPLESG-VSLLSRPVLVKKSATDLLPKWLWF 122
I F+ + ++ +S+L+ P P LF + G +S + L V + D++PK+L F
Sbjct 4 IHFTTEGEVTFKSILFVPTSDPRGLFDEYGSKKSDYIKLYVLCVFITDEFHDMMPKYLNF 63
Query 123 LRGIVDCEDLPLNVSREHMQDSALQRKLSSVIVRRIL 159
++G+VD +DLPLNVS E +Q Q KL VI ++++
Sbjct 64 VKGVVDSDDLPLNVSLETLQ----QHKLLKVIRKKLV 96
> ECU02g1100
Length=690
Score = 59.3 bits (142), Expect = 3e-09, Method: Composition-based stats.
Identities = 41/201 (20%), Positives = 82/201 (40%), Gaps = 40/201 (19%)
Query 2 RVKECAQKFSAFVNFPVYM---------------------------EDNGKEV------- 27
R+ E +K+S FV +P+Y E +EV
Sbjct 210 RISEIVKKYSLFVFYPIYTYVEKEIEEPEEKKDEEKEDEKVEEETAEPRVEEVREKRLKK 269
Query 28 -----QVSSQEALWLKT-SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYF 81
Q++ ++ LW + PEE + F + W + + + LSI +++
Sbjct 270 VTEREQINVEKPLWKRNIKEVPEEELKSFYKTVSGDWDDFLAVDFWHIEGLLSIELLMFI 329
Query 82 PADPPNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHM 141
P +F + + L + V V D +P+W+ F+ G+V +D+P+N+SRE +
Sbjct 330 PKRARFDMFNKNKKNNNIKLYCKNVFVTDDFGDAIPEWMSFVSGVVASDDIPMNISREMI 389
Query 142 QDSALQRKLSSVIVRRILRFL 162
Q + + + + + ++I +
Sbjct 390 QGTNVMKLVKKTLPQKIFEMI 410
> Hs20535880
Length=340
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 41/69 (59%), Gaps = 0/69 (0%)
Query 96 ESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSALQRKLSSVIV 155
++ + L V + + +L+P++L F+R +VD EDLPLN+S + +Q S + + + +V
Sbjct 207 KNNIKLYVCRVFITDNCEELIPEYLNFIRVLVDSEDLPLNISHKMLQQSKILKVIRKNLV 266
Query 156 RRILRFLND 164
++ L +
Sbjct 267 KKCLELFTE 275
> Hs18550811
Length=329
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 37/59 (62%), Gaps = 0/59 (0%)
Query 106 VLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSALQRKLSSVIVRRILRFLND 164
V + S +L ++L F+ G+VD + LPLN+SRE ++ S + + + + IV++ L ++
Sbjct 121 VFIMDSCDELTQEFLNFIHGLVDSQYLPLNISREMLKQSKILKVIQTNIVKKCLELFSE 179
> Hs20545837_2
Length=187
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 23/102 (22%), Positives = 47/102 (46%), Gaps = 2/102 (1%)
Query 28 QVSSQEALWLKT-SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPP 86
+++ + +W + +E+ +F TN W + FS + L R++L+ P P
Sbjct 32 ELNKIKTIWTRNHDYITQEYGEFCNSHTN-DWEDHLAVKHFSIEGQLEFRALLFIPYRAP 90
Query 87 NRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVD 128
LF+ + + L V + S +L+P+ L F+ I++
Sbjct 91 FDLFENNKNKDTIKLYVLHVFIVHSCDELIPEDLNFICAIME 132
> At1g54010
Length=426
Score = 31.6 bits (70), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 30/109 (27%), Positives = 45/109 (41%), Gaps = 19/109 (17%)
Query 8 QKFSAFVNFPVYMEDNGKEVQVSSQEALWLKTSATPEEHKQ---------FF--RHLTNQ 56
Q F VN+ + N V + +A P H + FF RH T +
Sbjct 279 QNFRELVNYHGFFVTNASCCGVGTHDAYG---CGFPNVHSRLCEYQRSYLFFDGRHNTEK 335
Query 57 SWGEPFYSIMFSADA----PLSIRSVLYFPADPPNRLFQTGPLESGVSL 101
+ E F ++F AD P++IR ++ +PAD P R P + V L
Sbjct 336 AQ-EMFGHLLFGADTNVIQPMNIRELVVYPADEPMRESWVPPTSATVQL 383
> Hs22065017
Length=343
Score = 31.2 bits (69), Expect = 0.89, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 24/41 (58%), Gaps = 1/41 (2%)
Query 42 TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFP 82
T EE+ +F+++LTN W + FS + L R++L+ P
Sbjct 304 TNEEYGEFYKNLTN-DWEDHLAVKHFSVEGQLEFRALLFVP 343
> Hs20534409
Length=1346
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 17/67 (25%), Positives = 27/67 (40%), Gaps = 0/67 (0%)
Query 37 LKTSATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPPNRLFQTGPLE 96
LK A ++ H+TN W F ++ + A S+ + P N + +T P E
Sbjct 436 LKRGAKFRKYVGHSAHVTNVRWSHDFQWVLSTGGADHSVFQWRFIPEGVSNGMLETAPQE 495
Query 97 SGVSLLS 103
G S
Sbjct 496 GGADSYS 502
> Hs5803066
Length=489
Score = 28.5 bits (62), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 22/34 (64%), Gaps = 5/34 (14%)
Query 53 LTNQSWGEPFYSIMFS----ADAPLSIRSVLYFP 82
L QSWG PF++ + + ADAPL +RS+ +P
Sbjct 344 LLCQSWG-PFHTFLLTKAGAADAPLRLRSIHEYP 376
> Hs20536809
Length=297
Score = 28.5 bits (62), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 25/48 (52%), Gaps = 9/48 (18%)
Query 72 PLSIRSVLYFPADPPNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKW 119
P++I +L++P+ PP+ S LLSRP K+ + +L W
Sbjct 205 PVTIHCLLHYPSSPPH---------STAQLLSRPSGGKEQSPELQQDW 243
> CE22992
Length=541
Score = 28.5 bits (62), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 19/45 (42%), Positives = 26/45 (57%), Gaps = 4/45 (8%)
Query 122 FLRGIVDCEDLPLNVSREHM---QDSALQRKLSSVIVRRILRFLN 163
F+R +VD E VSRE M D A Q+ SS+ R+++FLN
Sbjct 131 FIRFLVD-EQQTTTVSRELMIRSNDVATQQVFSSIRPSRVIKFLN 174
Lambda K H
0.321 0.135 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2353551590
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40