bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
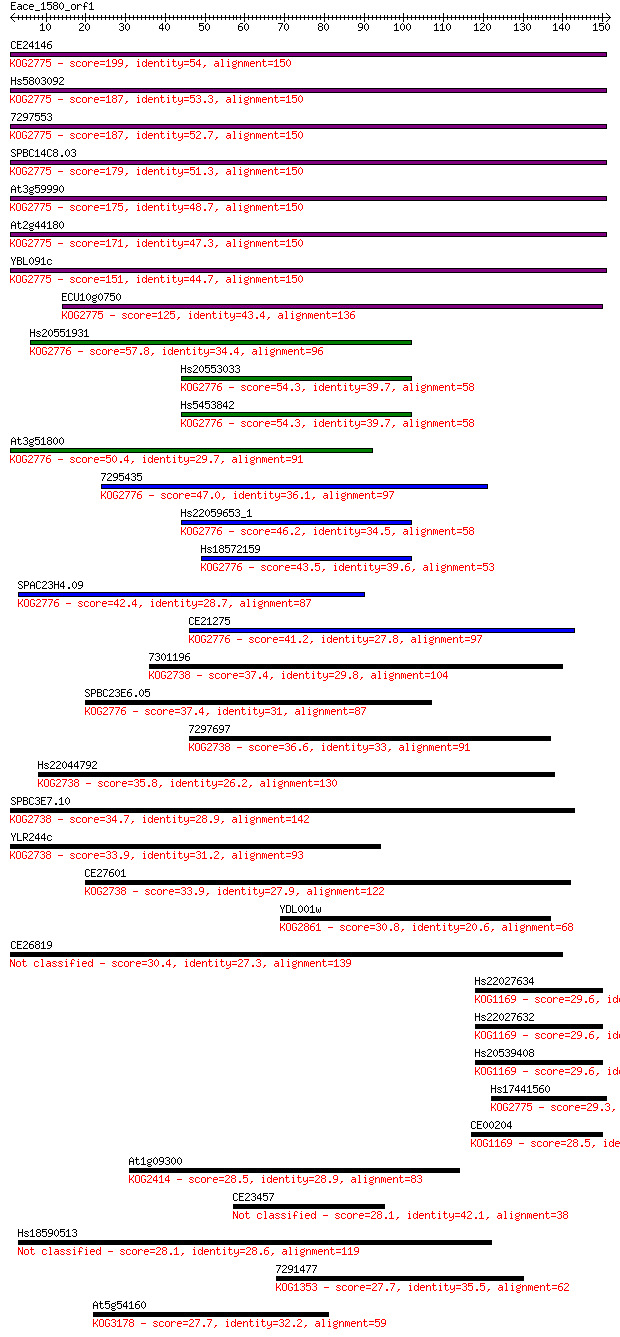
Query= Eace_1580_orf1
Length=151
Score E
Sequences producing significant alignments: (Bits) Value
CE24146 199 2e-51
Hs5803092 187 5e-48
7297553 187 1e-47
SPBC14C8.03 179 2e-45
At3g59990 175 2e-44
At2g44180 171 3e-43
YBL091c 151 4e-37
ECU10g0750 125 3e-29
Hs20551931 57.8 8e-09
Hs20553033 54.3 9e-08
Hs5453842 54.3 1e-07
At3g51800 50.4 1e-06
7295435 47.0 1e-05
Hs22059653_1 46.2 2e-05
Hs18572159 43.5 1e-04
SPAC23H4.09 42.4 3e-04
CE21275 41.2 7e-04
7301196 37.4 0.010
SPBC23E6.05 37.4 0.011
7297697 36.6 0.020
Hs22044792 35.8 0.034
SPBC3E7.10 34.7 0.080
YLR244c 33.9 0.11
CE27601 33.9 0.12
YDL001w 30.8 1.1
CE26819 30.4 1.3
Hs22027634 29.6 2.0
Hs22027632 29.6 2.0
Hs20539408 29.6 2.1
Hs17441560 29.3 2.6
CE00204 28.5 5.4
At1g09300 28.5 5.4
CE23457 28.1 6.7
Hs18590513 28.1 6.8
7291477 27.7 7.7
At5g54160 27.7 9.1
> CE24146
Length=444
Score = 199 bits (505), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 81/150 (54%), Positives = 117/150 (78%), Gaps = 0/150 (0%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECA 60
R +AE HRQVR+Y +S ++PG+++ E+C +LE + LI G+E G FPTGCSLN CA
Sbjct 134 RRSAEAHRQVRKYVKSWIKPGMTMIEICERLETTSRRLIKEQGLEAGLAFPTGCSLNHCA 193
Query 61 AHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLR 120
AHYTPN G+ +L GD+CK+D+G+ VRGR+ID AF++ FDPKFDPL++A +EATNAG++
Sbjct 194 AHYTPNAGDTTVLQYGDVCKIDYGIHVRGRLIDSAFTVHFDPKFDPLVEAVREATNAGIK 253
Query 121 AAGVDARLSELGCIIEETINSFEMQLDGQT 150
+G+D RL ++G I+EE + S E++LDG++
Sbjct 254 ESGIDVRLCDVGEIVEEVMTSHEVELDGKS 283
> Hs5803092
Length=478
Score = 187 bits (476), Expect = 5e-48, Method: Compositional matrix adjust.
Identities = 80/150 (53%), Positives = 113/150 (75%), Gaps = 0/150 (0%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECA 60
REAAE HRQVR+Y S ++PG+++ E+C KLE + +LI G+ G FPTGCSLN CA
Sbjct 170 REAAEAHRQVRKYVMSWIKPGMTMIEICEKLEDCSRKLIKENGLNAGLAFPTGCSLNNCA 229
Query 61 AHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLR 120
AHYTPN G+ +L DICK+DFG + GRIIDCAF++ F+PK+D L++A K+ATN G++
Sbjct 230 AHYTPNAGDTTVLQYDDICKIDFGTHISGRIIDCAFTVTFNPKYDTLLKAVKDATNTGIK 289
Query 121 AAGVDARLSELGCIIEETINSFEMQLDGQT 150
AG+D RL ++G I+E + S+E+++DG+T
Sbjct 290 CAGIDVRLCDVGEAIQEVMESYEVEIDGKT 319
> 7297553
Length=448
Score = 187 bits (474), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 79/150 (52%), Positives = 111/150 (74%), Gaps = 0/150 (0%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECA 60
R+AAE HRQ R+Y Q ++PG+++ ++C +LE LI G+E G FPTGCSLN CA
Sbjct 140 RQAAEAHRQTRQYMQRYIKPGMTMIQICEELENTARRLIGENGLEAGLAFPTGCSLNHCA 199
Query 61 AHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLR 120
AHYTPN G+ +L D+CK+DFG ++GRIIDCAF++ F+ K+D L+QA KEATN G+R
Sbjct 200 AHYTPNAGDPTVLQYDDVCKIDFGTHIKGRIIDCAFTLTFNNKYDKLLQAVKEATNTGIR 259
Query 121 AAGVDARLSELGCIIEETINSFEMQLDGQT 150
AG+D RL ++G I+E + S+E++LDG+T
Sbjct 260 EAGIDVRLCDIGAAIQEVMESYEIELDGKT 289
> SPBC14C8.03
Length=426
Score = 179 bits (454), Expect = 2e-45, Method: Compositional matrix adjust.
Identities = 77/150 (51%), Positives = 112/150 (74%), Gaps = 0/150 (0%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECA 60
R AAE HRQ R+YAQS+++PG+S+ ++ N +E T L+ G++ G GFPTG SLN CA
Sbjct 118 RRAAEVHRQARQYAQSVIKPGMSMMDVVNTIENTTRALVEEDGLKSGIGFPTGVSLNHCA 177
Query 61 AHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLR 120
AHYTPN G+ IL + D+ K+D GV V GRI+D AF+++FDP++D L+ A K ATN G+
Sbjct 178 AHYTPNAGDTTILKEKDVMKVDIGVHVNGRIVDSAFTMSFDPQYDNLLAAVKAATNKGIE 237
Query 121 AAGVDARLSELGCIIEETINSFEMQLDGQT 150
AG+DARL+E+G I+E + S+E++++G+T
Sbjct 238 EAGIDARLNEIGEAIQEVMESYEVEINGKT 267
> At3g59990
Length=435
Score = 175 bits (444), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 73/150 (48%), Positives = 110/150 (73%), Gaps = 0/150 (0%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECA 60
R AAE HRQVR+Y +S+++PG+ +T++C LE +LI+ G++ G FPTGCSLN A
Sbjct 127 RRAAEVHRQVRKYVRSIVKPGMLMTDICETLENTVRKLISENGLQAGIAFPTGCSLNWVA 186
Query 61 AHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLR 120
AH+TPN G+ +L D+ KLDFG + G IIDCAF++AF+P FDPL+ A++EAT G++
Sbjct 187 AHWTPNSGDKTVLQYDDVMKLDFGTHIDGHIIDCAFTVAFNPMFDPLLAASREATYTGIK 246
Query 121 AAGVDARLSELGCIIEETINSFEMQLDGQT 150
AG+D RL ++G I+E + S+E++++G+
Sbjct 247 EAGIDVRLCDIGAAIQEVMESYEVEINGKV 276
> At2g44180
Length=431
Score = 171 bits (434), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 71/150 (47%), Positives = 109/150 (72%), Gaps = 0/150 (0%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECA 60
R+AAE HRQVR+Y +S+L+PG+ + +LC LE +LI+ G++ G FPTGCSLN A
Sbjct 123 RQAAEVHRQVRKYMRSILKPGMLMIDLCETLENTVRKLISENGLQAGIAFPTGCSLNNVA 182
Query 61 AHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLR 120
AH+TPN G+ +L D+ KLDFG + G I+D AF++AF+P FDPL+ A+++AT G++
Sbjct 183 AHWTPNSGDKTVLQYDDVMKLDFGTHIDGHIVDSAFTVAFNPMFDPLLAASRDATYTGIK 242
Query 121 AAGVDARLSELGCIIEETINSFEMQLDGQT 150
AGVD RL ++G ++E + S+E++++G+
Sbjct 243 EAGVDVRLCDVGAAVQEVMESYEVEINGKV 272
> YBL091c
Length=421
Score = 151 bits (382), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 67/157 (42%), Positives = 106/157 (67%), Gaps = 7/157 (4%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGI-------ERGKGFPTG 53
R+ AE HR+VRR + + PG+ L ++ + +E T + A + +G GFPTG
Sbjct 106 RKGAEIHRRVRRAIKDRIVPGMKLMDIADMIENTTRKYTGAENLLAMEDPKSQGIGFPTG 165
Query 54 CSLNECAAHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKE 113
SLN CAAH+TPN G+ +L D+ K+D+GVQV G IID AF+++FDP++D L+ A K+
Sbjct 166 LSLNHCAAHFTPNAGDKTVLKYEDVMKVDYGVQVNGNIIDSAFTVSFDPQYDNLLAAVKD 225
Query 114 ATNAGLRAAGVDARLSELGCIIEETINSFEMQLDGQT 150
AT G++ AG+D RL+++G I+E + S+E++++G+T
Sbjct 226 ATYTGIKEAGIDVRLTDIGEAIQEVMESYEVEINGET 262
> ECU10g0750
Length=358
Score = 125 bits (314), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 59/137 (43%), Positives = 89/137 (64%), Gaps = 2/137 (1%)
Query 14 AQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECAAHYTPNPGE-DRI 72
QS++RPG++L E+ +E T L+ G GFP G S+N CAAHYT NPGE D +
Sbjct 62 VQSIVRPGITLLEIVRSIEDSTRTLLKGER-NNGIGFPAGMSMNSCAAHYTVNPGEQDIV 120
Query 73 LGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLRAAGVDARLSELG 132
L + D+ K+DFG GRI+D AF++AF +PL+ A +E T G+++ GVD R+ ++G
Sbjct 121 LKEDDVLKIDFGTHSDGRIMDSAFTVAFKENLEPLLVAAREGTETGIKSLGVDVRVCDIG 180
Query 133 CIIEETINSFEMQLDGQ 149
I E I+S+E+++ G+
Sbjct 181 RDINEVISSYEVEIGGR 197
> Hs20551931
Length=307
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 33/102 (32%), Positives = 51/102 (50%), Gaps = 7/102 (6%)
Query 6 CHRQVRRYAQSLLRPGLSLTELCNKLEAKTEE-----LIAAAGIERGKGFPTGCSLNECA 60
HR +R ++ G+ + LC K +A E +++G FPT S+N C
Sbjct 6 AHRVLRSLVEAS-SSGVLVLSLCEKSDAVIMEETGKIFKKEKEMKKGIAFPTSISVNNCV 64
Query 61 AHYTP-NPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFD 101
H++P G+D IL +GD+ K+D GV V G I + A + D
Sbjct 65 CHFSPLESGQDYILKEGDLVKIDLGVYVDGFIANVAHTFVVD 106
> Hs20553033
Length=210
Score = 54.3 bits (129), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 23/59 (38%), Positives = 35/59 (59%), Gaps = 1/59 (1%)
Query 44 IERGKGFPTGCSLNECAAHYTP-NPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFD 101
+++G FPT S+N C H++P +D IL +GD+ K+D GV V G I + A + D
Sbjct 70 MKKGIAFPTSISVNNCVCHFSPLKSDQDYILKEGDLVKIDLGVHVDGFIANVAHTFVVD 128
> Hs5453842
Length=394
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 23/59 (38%), Positives = 35/59 (59%), Gaps = 1/59 (1%)
Query 44 IERGKGFPTGCSLNECAAHYTP-NPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFD 101
+++G FPT S+N C H++P +D IL +GD+ K+D GV V G I + A + D
Sbjct 70 MKKGIAFPTSISVNNCVCHFSPLKSDQDYILKEGDLVKIDLGVHVDGFIANVAHTFVVD 128
> At3g51800
Length=392
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 48/97 (49%), Gaps = 6/97 (6%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAA------AGIERGKGFPTGC 54
+ AAE + + + +P + ++C K ++ +E A+ IERG FPT
Sbjct 24 KSAAEIVNKALQVVLAECKPKAKIVDICEKGDSFIKEQTASMYKNSKKKIERGVAFPTCI 83
Query 55 SLNECAAHYTPNPGEDRILGQGDICKLDFGVQVRGRI 91
S+N H++P ++ +L GD+ K+D G + G I
Sbjct 84 SVNNTVGHFSPLASDESVLEDGDMVKIDMGCHIDGFI 120
> 7295435
Length=391
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 35/106 (33%), Positives = 49/106 (46%), Gaps = 15/106 (14%)
Query 24 LTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECAAHYTPNPGE-DRILGQGDICKLD 82
LTE K+ K ++L ++G FPT S+N C H++P + D L GD+ K+D
Sbjct 56 LTEETGKVYKKEKDL------KKGIAFPTCLSVNNCVCHFSPAKNDADYTLKAGDVVKID 109
Query 83 FGVQVRGRIIDCAFSI----AFDPKFD----PLIQATKEATNAGLR 120
G + G I A +I A D K +I A A A LR
Sbjct 110 LGAHIDGFIAVAAHTIVVGAAADQKISGRQADVILAAYWAVQAALR 155
> Hs22059653_1
Length=215
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 20/59 (33%), Positives = 33/59 (55%), Gaps = 1/59 (1%)
Query 44 IERGKGFPTGCSLNECAAHYTP-NPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFD 101
+++ FPT +N C H++P +D IL +G++ K+D GVQV G I + + D
Sbjct 35 MKKSIAFPTSILVNNCVGHFSPLKSDKDCILKEGNLVKIDLGVQVDGFIANVTHTFVVD 93
> Hs18572159
Length=910
Score = 43.5 bits (101), Expect = 1e-04, Method: Composition-based stats.
Identities = 21/54 (38%), Positives = 30/54 (55%), Gaps = 1/54 (1%)
Query 49 GFPTGCSLNECAAHYTP-NPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFD 101
FPT +N C H++P +D IL +GD+ K+D GV V G I + A + D
Sbjct 483 AFPTSILVNNCVCHFSPLKRDQDYILKEGDLVKIDLGVHVNGFITNVARTFLTD 536
> SPAC23H4.09
Length=381
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/94 (26%), Positives = 43/94 (45%), Gaps = 7/94 (7%)
Query 3 AAECHRQVRRYAQSLLRPGLSLTELCNK----LEAKTEELIAAAGIERGKGFPTGCSLNE 58
A E + V + L +PG + ++C + L +++ +G FPT S N+
Sbjct 28 AGEVSQNVIKKVVELCQPGAKIYDICVRGDELLNEAIKKVYRTKDAYKGIAFPTAVSPND 87
Query 59 CAAHYTP---NPGEDRILGQGDICKLDFGVQVRG 89
AAH +P +P + L GD+ K+ G + G
Sbjct 88 MAAHLSPLKSDPEANLALKSGDVVKILLGAHIDG 121
> CE21275
Length=391
Score = 41.2 bits (95), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 27/106 (25%), Positives = 45/106 (42%), Gaps = 9/106 (8%)
Query 46 RGKGFPTGCSLNECAAHYTPNPGE-DRILGQGDICKLDFGVQVRGRIIDCAFSIAFDP-- 102
+G PT S++ C HYTP E +L G + K+D G + G I A ++
Sbjct 83 KGIAMPTCISIDNCICHYTPLKSEAPVVLKNGQVVKVDLGTHIDGLIATAAHTVVVGASK 142
Query 103 ------KFDPLIQATKEATNAGLRAAGVDARLSELGCIIEETINSF 142
K L++ T +A +R+ D + + I++T F
Sbjct 143 DNKVTGKLADLLRGTHDALEIAIRSLRPDTENTTITKNIDKTAAEF 188
> 7301196
Length=374
Score = 37.4 bits (85), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 31/117 (26%), Positives = 49/117 (41%), Gaps = 16/117 (13%)
Query 36 EELIAAAGIER--------GKGFPTGC--SLNECAAHYTPNPGEDRILGQGDICKLDFGV 85
+ L+ A IER FP C S+NE H P + R L GD+C +D V
Sbjct 154 DRLVHEAAIERECYPSPLNYYNFPKSCCTSVNEVICHGIP---DQRPLQDGDLCNIDVTV 210
Query 86 QVR---GRIIDCAFSIAFDPKFDPLIQATKEATNAGLRAAGVDARLSELGCIIEETI 139
R G + + F K L+Q T EA + + + ++G +I++ +
Sbjct 211 YHRGFHGDLNETFFVGNVSEKHKKLVQVTHEALSKAIEFVRPGEKYRDIGNVIQKYV 267
> SPBC23E6.05
Length=417
Score = 37.4 bits (85), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 27/98 (27%), Positives = 38/98 (38%), Gaps = 11/98 (11%)
Query 20 PGLSLTELC----NKLEAKTEELIAAAGIERGKGFPTGCSLNECAAHYTPNP-----GED 70
PG S E+ N L + + E+G PT +N CA +Y P P G D
Sbjct 41 PGASTREISSYGDNLLHEYKSSIYKSQRFEKGIAEPTSICVNNCAYNYAPGPESVIAGND 100
Query 71 RI--LGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDP 106
L GD+ K+ G+ G + +I P P
Sbjct 101 NSYHLQVGDVTKISMGLHFDGYTALISHTIVVTPPPQP 138
> 7297697
Length=307
Score = 36.6 bits (83), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 30/96 (31%), Positives = 44/96 (45%), Gaps = 8/96 (8%)
Query 46 RGKGFPTG--CSLNECAAHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCA--FSIA-F 100
R GFP S+N A H P +DR L GDI +D V + G DC+ F +
Sbjct 118 RYAGFPKSICTSINNIACHGIP---DDRQLADGDIINIDVTVFLNGYHGDCSETFRVGNV 174
Query 101 DPKFDPLIQATKEATNAGLRAAGVDARLSELGCIIE 136
D + L++ATK + + G +E+G I+
Sbjct 175 DERGGFLVEATKSCLDQCISLCGPGVEFNEIGKFID 210
> Hs22044792
Length=386
Score = 35.8 bits (81), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 34/135 (25%), Positives = 54/135 (40%), Gaps = 8/135 (5%)
Query 8 RQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGC--SLNECAAHYTP 65
R+V A +++PG++ E+ + + FP C S+NE H P
Sbjct 147 REVLDVAAGMIKPGVTTEEIDHAVHLACIARNCYPSPLNYYNFPKSCCTSVNEVICHGIP 206
Query 66 NPGEDRILGQGDICKLDFGVQ---VRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLRAA 122
+ R L +GDI +D + G + + F D L+Q T E + A
Sbjct 207 ---DRRPLQEGDIVNVDITLYRNGYHGDLNETFFVGEVDDGARKLVQTTYECLMQAIDAV 263
Query 123 GVDARLSELGCIIEE 137
R ELG II++
Sbjct 264 KPGVRYRELGNIIQK 278
> SPBC3E7.10
Length=379
Score = 34.7 bits (78), Expect = 0.080, Method: Compositional matrix adjust.
Identities = 41/158 (25%), Positives = 62/158 (39%), Gaps = 27/158 (17%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERG--------KGFPT 52
R+ R+V A + +RPG + EL + ++ A IER FP
Sbjct 129 RKVCRLGREVLDAAAAAVRPGTTTDEL--------DSIVHNACIERDCFPSTLNYYAFPK 180
Query 53 GC--SLNECAAHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDC--AFSIAFDPKFDP-- 106
S+NE H P + R L GDI +D + G D + + K +P
Sbjct 181 SVCTSVNEIICHGIP---DQRPLEDGDIVNIDVSLYHNGFHGDLNETYYVGDKAKANPDL 237
Query 107 --LIQATKEATNAGLRAAGVDARLSELGCIIEETINSF 142
L++ T+ A + + A E G IIE+ NS
Sbjct 238 VCLVENTRIALDKAIAAVKPGVLFQEFGNIIEKHTNSI 275
> YLR244c
Length=387
Score = 33.9 bits (76), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 29/103 (28%), Positives = 45/103 (43%), Gaps = 21/103 (20%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERG--------KGFPT 52
R+A R+V A + +RPG++ EL +E++ I+RG FP
Sbjct 139 RKACMLGREVLDIAAAHVRPGITTDEL--------DEIVHNETIKRGAYPSPLNYYNFPK 190
Query 53 G--CSLNECAAHYTPNPGEDRILGQGDICKLDFGVQVRGRIID 93
S+NE H P + +L +GDI LD + +G D
Sbjct 191 SLCTSVNEVICHGVP---DKTVLKEGDIVNLDVSLYYQGYHAD 230
> CE27601
Length=388
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 34/136 (25%), Positives = 54/136 (39%), Gaps = 25/136 (18%)
Query 20 PGLSLTELCNKLEAKTEELIAAAGIERG--------KGFPTGC--SLNECAAHYTPNPGE 69
PG++ E+ + ++ A IER FP C S+NE H P +
Sbjct 140 PGVTTEEI--------DRVVHEAAIERDCYPSPLGYYKFPKSCCTSVNEVICHGIP---D 188
Query 70 DRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFD----PLIQATKEATNAGLRAAGVD 125
R L GD+C +D V RG D + K D L++ T E +
Sbjct 189 MRKLENGDLCNVDVTVYHRGFHGDLNETFLVGDKVDEESRKLVKVTFECLQQAIAIVKPG 248
Query 126 ARLSELGCIIEETINS 141
+ E+G +I++ N+
Sbjct 249 VKFREIGNVIQKHANA 264
> YDL001w
Length=430
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 14/68 (20%), Positives = 34/68 (50%), Gaps = 0/68 (0%)
Query 69 EDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLRAAGVDARL 128
ED + G++ L + + G ++D + +P+ +P+ QAT+ R + ++ RL
Sbjct 321 EDIMKSIGELFILRININLHGSVLDSPEIMWSEPQLEPIYQATRGYLEINQRVSLLNQRL 380
Query 129 SELGCIIE 136
+ +++
Sbjct 381 EVISDLLQ 388
> CE26819
Length=498
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 38/152 (25%), Positives = 55/152 (36%), Gaps = 23/152 (15%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKG--------FPT 52
R A++ + R A +RPGL E + E L G T
Sbjct 193 RYASKIASEAHRAAMKHMRPGL--------YEYQLESLFRHTSYYHGGCRHLAYTCIAAT 244
Query 53 GCSLNECAAHYT-PNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDP----L 107
GC N HY N D+ + GD+C D G + D S + KF +
Sbjct 245 GC--NGSVLHYGHANAPNDKFIKDGDMCLFDMGPEYNCYASDITTSFPSNGKFTEKQKIV 302
Query 108 IQATKEATNAGLRAAGVDARLSELGCIIEETI 139
A A A L+AA R +++ + E+ I
Sbjct 303 YNAVLAANLAVLKAAKPGVRWTDMHILSEKVI 334
> Hs22027634
Length=773
Score = 29.6 bits (65), Expect = 2.0, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 21/32 (65%), Gaps = 2/32 (6%)
Query 118 GLRAAGVDARLSELGCIIEETINSFEMQLDGQ 149
GL++AG RL++ C++ T S MQ+DG+
Sbjct 732 GLKSAG--RRLAQCSCVVIRTSKSLPMQIDGE 761
> Hs22027632
Length=804
Score = 29.6 bits (65), Expect = 2.0, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 21/32 (65%), Gaps = 2/32 (6%)
Query 118 GLRAAGVDARLSELGCIIEETINSFEMQLDGQ 149
GL++AG RL++ C++ T S MQ+DG+
Sbjct 732 GLKSAG--RRLAQCSCVVIRTSKSLPMQIDGE 761
> Hs20539408
Length=738
Score = 29.6 bits (65), Expect = 2.1, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 21/32 (65%), Gaps = 2/32 (6%)
Query 118 GLRAAGVDARLSELGCIIEETINSFEMQLDGQ 149
GL++AG RL++ C++ T S MQ+DG+
Sbjct 666 GLKSAG--RRLAQCSCVVIRTSKSLPMQIDGE 695
> Hs17441560
Length=200
Score = 29.3 bits (64), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 11/29 (37%), Positives = 22/29 (75%), Gaps = 0/29 (0%)
Query 122 AGVDARLSELGCIIEETINSFEMQLDGQT 150
AG+D L ++G I++ + S+E+++DG+T
Sbjct 90 AGIDVHLCDVGESIQKFMESYEVEIDGKT 118
> CE00204
Length=827
Score = 28.5 bits (62), Expect = 5.4, Method: Composition-based stats.
Identities = 14/33 (42%), Positives = 21/33 (63%), Gaps = 3/33 (9%)
Query 117 AGLRAAGVDARLSELGCIIEETINSFEMQLDGQ 149
AG+R A RLS+ ++ +T SF MQ+DG+
Sbjct 738 AGVRGA---RRLSQCSTVVIQTHKSFPMQIDGE 767
> At1g09300
Length=451
Score = 28.5 bits (62), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 24/92 (26%), Positives = 38/92 (41%), Gaps = 12/92 (13%)
Query 31 LEAKTEELIAAAGIERGKGFPT-GCSLNECAAHYTPNPGEDRILGQGDICKLDFGVQVRG 89
L A+ E G +R P G N HY+ N D+ + GD+ +D G ++ G
Sbjct 205 LSAQVEYECRVRGAQRMAFNPVVGGGSNASVIHYSRN---DQRIKDGDLVLMDMGCELHG 261
Query 90 RIIDCA--------FSIAFDPKFDPLIQATKE 113
+ D FS + +D ++Q KE
Sbjct 262 YVSDLTRTWPPCGKFSSVQEELYDLILQTNKE 293
> CE23457
Length=485
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 20/40 (50%), Gaps = 3/40 (7%)
Query 57 NECAAHYTPNPGED--RILGQGDICKLDFGVQVRGRIIDC 94
NECA H T NP D + QG + + F + V G DC
Sbjct 155 NECAFHVTDNPKLDATHLCAQGAVADM-FDMIVTGNFNDC 193
> Hs18590513
Length=493
Score = 28.1 bits (61), Expect = 6.8, Method: Composition-based stats.
Identities = 34/125 (27%), Positives = 50/125 (40%), Gaps = 17/125 (13%)
Query 3 AAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECAA- 61
++E HR+V + ++ G+ EL E+ E + G R + C E +A
Sbjct 202 SSEAHREVMK----AVKVGMKEYEL----ESLFEHYCYSRGGMRHSSYTCICGSGENSAV 253
Query 62 -HY----TPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATN 116
HY PN DR + GD+C D G + D S + KF +A EA
Sbjct 254 LHYGHAGAPN---DRTIQNGDMCLFDMGGEYYCFASDITCSFPANGKFTADQKAVYEAVL 310
Query 117 AGLRA 121
RA
Sbjct 311 RSSRA 315
> 7291477
Length=552
Score = 27.7 bits (60), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 35/69 (50%), Gaps = 11/69 (15%)
Query 68 GEDRILGQGDICK-------LDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLR 120
G D+++ QGDI K + G ++ GR++D A A D K TK+ G++
Sbjct 119 GNDKLIKQGDIVKRTGAIVDVPVGDELLGRVVD-ALGNAIDGKGAI---NTKDRFRVGIK 174
Query 121 AAGVDARLS 129
A G+ R+S
Sbjct 175 APGIIPRVS 183
> At5g54160
Length=363
Score = 27.7 bits (60), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 28/59 (47%), Gaps = 2/59 (3%)
Query 22 LSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECAAHYTPNPGEDRILGQGDICK 80
+S TE+ +KL K E A ++R T S+ C+ G +RI G G +CK
Sbjct 55 MSPTEIASKLPTKNPE--APVMLDRILRLLTSYSVLTCSNRKLSGDGVERIYGLGPVCK 111
Lambda K H
0.320 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1888713112
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40