bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1570_orf1
Length=128
Score E
Sequences producing significant alignments: (Bits) Value
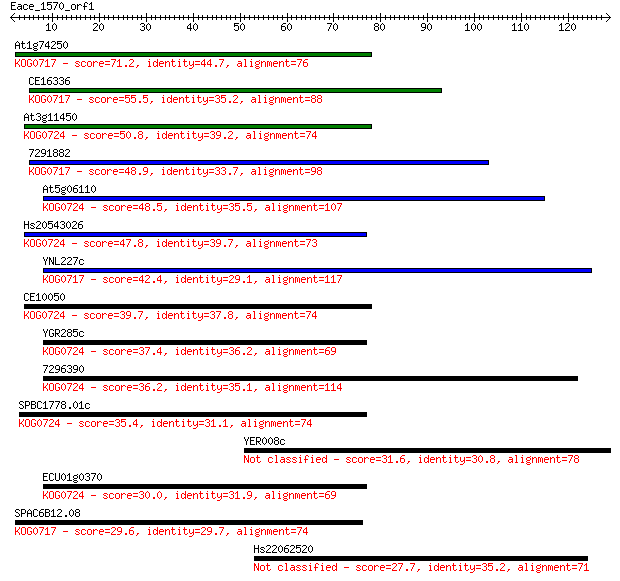
At1g74250 71.2 4e-13
CE16336 55.5 3e-08
At3g11450 50.8 7e-07
7291882 48.9 2e-06
At5g06110 48.5 3e-06
Hs20543026 47.8 5e-06
YNL227c 42.4 2e-04
CE10050 39.7 0.001
YGR285c 37.4 0.006
7296390 36.2 0.016
SPBC1778.01c 35.4 0.028
YER008c 31.6 0.34
ECU01g0370 30.0 1.2
SPAC6B12.08 29.6 1.6
Hs22062520 27.7 5.6
> At1g74250
Length=630
Score = 71.2 bits (173), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 34/76 (44%), Positives = 50/76 (65%), Gaps = 2/76 (2%)
Query 2 WGDVGPFYSYWHSFTSVRDFAEVDQWSPKDMAEASRAERRLMERDNGRKRKEAKKEFVDT 61
+ V FY+YW F +V DF VD++ M +R RR+ME +N + RK+AK+E+ DT
Sbjct 165 YAQVTAFYNYWLGFCTVMDFCWVDEYDV--MGGPNRKSRRMMEEENKKSRKKAKREYNDT 222
Query 62 VRRLAQHIKKKDPRVL 77
VR LA+ +KK+D RV+
Sbjct 223 VRGLAEFVKKRDKRVI 238
> CE16336
Length=510
Score = 55.5 bits (132), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 31/89 (34%), Positives = 54/89 (60%), Gaps = 4/89 (4%)
Query 5 VGPFYSYWHSFTSVRDFAEVDQWSPKDMAEAS-RAERRLMERDNGRKRKEAKKEFVDTVR 63
V FY +W SF++ R FA +D + D+ +AS R E R ++++N + R K+E + +R
Sbjct 171 VNGFYGFWSSFSTTRSFAWLDHY---DITQASNRFESRQIDQENKKFRDVGKQERNEQIR 227
Query 64 RLAQHIKKKDPRVLARQAALAQQRLDEQR 92
L ++K+DPRV A + L Q++L+ +
Sbjct 228 NLVAFVRKRDPRVKAYREILEQKKLEAHK 256
> At3g11450
Length=663
Score = 50.8 bits (120), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 29/75 (38%), Positives = 49/75 (65%), Gaps = 4/75 (5%)
Query 4 DVGPFYSYWHSFTSVRDFAEVDQWSPKDMAEA-SRAERRLMERDNGRKRKEAKKEFVDTV 62
DV FY++W++F S R+F + ++ D+ +A SR ERR ME++N +K +A+KE +
Sbjct 244 DVDKFYNFWYAFKSWREFPDEEE---HDLEQADSREERRWMEKENAKKTVKARKEEHARI 300
Query 63 RRLAQHIKKKDPRVL 77
R L + +KDPR++
Sbjct 301 RTLVDNAYRKDPRIV 315
> 7291882
Length=540
Score = 48.9 bits (115), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/99 (33%), Positives = 61/99 (61%), Gaps = 5/99 (5%)
Query 5 VGPFYSYWHSFTSVRDFAEVDQWSPKDMAEAS-RAERRLMERDNGRKRKEAKKEFVDTVR 63
VGPFY++W ++++ + + D P D+ E R R +E++ + + A+KE + VR
Sbjct 148 VGPFYAFWQAYSTRKTY---DWLCPYDVREIKERFILRKVEKEMKKIVQAARKERNEEVR 204
Query 64 RLAQHIKKKDPRVLARQAALAQQRLDEQRLKEEEQQRVQ 102
L ++K+DPRV A + L ++R++ RLK+EE+++ Q
Sbjct 205 NLVNFVRKRDPRVQAYRRML-EERVEANRLKQEEKRKEQ 242
> At5g06110
Length=663
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 38/108 (35%), Positives = 64/108 (59%), Gaps = 17/108 (15%)
Query 8 FYSYWHSFTSVRDFAEVDQWSPKDMAEA-SRAERRLMERDNGRKRKEAKKEFVDTVRRLA 66
FYS W++F S R+F E ++ D+ +A SR E+R MER+N RK ++A+KE +R L
Sbjct 232 FYSTWYTFKSWREFPEEEE---HDIEQAESREEKRWMERENARKTQKARKEEYARIRTLV 288
Query 67 QHIKKKDPRVLARQAALAQQRLDEQRLKEEEQQRVQEQQQLLRQQRQQ 114
+ KKD R+ Q+R D+++ K ++Q+++ + +RQQ
Sbjct 289 DNAYKKDIRI--------QKRKDDEKAK-----KLQKKEAKVMAKRQQ 323
> Hs20543026
Length=621
Score = 47.8 bits (112), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 29/73 (39%), Positives = 45/73 (61%), Gaps = 2/73 (2%)
Query 4 DVGPFYSYWHSFTSVRDFAEVDQWSPKDMAEASRAERRLMERDNGRKRKEAKKEFVDTVR 63
DV FYS+W++F S R+F+ +D+ K+ AE R ERR +E+ N R + KKE ++ +R
Sbjct 210 DVDIFYSFWYNFDSWREFSYLDE-EEKEKAEC-RDERRWIEKQNRATRAQRKKEEMNRIR 267
Query 64 RLAQHIKKKDPRV 76
L + DPR+
Sbjct 268 TLVDNAYSCDPRI 280
> YNL227c
Length=590
Score = 42.4 bits (98), Expect = 2e-04, Method: Composition-based stats.
Identities = 34/124 (27%), Positives = 62/124 (50%), Gaps = 9/124 (7%)
Query 8 FYSYWHSFTSVRDFAEVDQWSPKDMAEASRAERRLMERDNGRKRKEAKKEFVDTVRRLAQ 67
FY W +F +++ F+ D++ R +R + R N + R++A+ E+ TV+R
Sbjct 194 FYKTWSAFNTLKSFSWKDEYMYSK--NYDRRTKREVNRRNEKARQQARNEYNKTVKRFVV 251
Query 68 HIKKKDPRVL-ARQAALAQQRLDEQRLKEEEQQRVQ------EQQQLLRQQRQQQQEEYY 120
IKK D R+ + A Q++L EQ+ K E R + ++++ Q Q +EE +
Sbjct 252 FIKKLDKRMKEGAKIAEEQRKLKEQQRKNELNNRRKFGNDNNDEEKFHLQSWQTVKEENW 311
Query 121 AALK 124
L+
Sbjct 312 DELE 315
> CE10050
Length=589
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 28/74 (37%), Positives = 43/74 (58%), Gaps = 2/74 (2%)
Query 4 DVGPFYSYWHSFTSVRDFAEVDQWSPKDMAEASRAERRLMERDNGRKRKEAKKEFVDTVR 63
DV FY +W +F S R+F+ +D+ K+ E R ERR ME+ N +R+ +KE +R
Sbjct 220 DVENFYDFWFNFQSWREFSYLDE-EDKERGE-DRYERREMEKQNKAERERRRKEEAKRIR 277
Query 64 RLAQHIKKKDPRVL 77
+L KDPR++
Sbjct 278 KLVDIAYAKDPRII 291
> YGR285c
Length=433
Score = 37.4 bits (85), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 38/71 (53%), Gaps = 6/71 (8%)
Query 8 FYSYWHSFTSVRDFAEVDQWSPKDMAEASRAERRLMERDNGRKRKEAKKEFVDTVR--RL 65
FY++WH F S R F +D+ P D ++R +R +ER N R KK+ D R +L
Sbjct 220 FYAFWHRFDSWRTFEFLDEDVPDD--SSNRDHKRYIERKNKAARD--KKKTADNARLVKL 275
Query 66 AQHIKKKDPRV 76
+ +DPR+
Sbjct 276 VERAVSEDPRI 286
> 7296390
Length=646
Score = 36.2 bits (82), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 40/152 (26%), Positives = 72/152 (47%), Gaps = 40/152 (26%)
Query 8 FYSYWHSFTSVRDFAEVDQWSPKDMAEASRAERRLMERDNGRKRKEAKKEFVDTVRRLAQ 67
FY++W+ F S R+F+ +D+ K+ + R ERR +E++N R + KKE + +R L
Sbjct 201 FYNFWYDFKSWREFSYLDE-EDKEKGQ-DRDERRWIEKENRAARIKRKKEEMSRIRSLVD 258
Query 68 HIKKKDPRV-------------------------------LARQAALAQQRLDEQRLKEE 96
D R+ R+AALA+++ ++ K
Sbjct 259 LAYNNDKRIQRFKQEEKDRKAAAKRAKMDAAQAQKAEADRAIREAALAKEKAEKAEQKRI 318
Query 97 EQQRVQEQQQ--LLRQQRQQQQE-----EYYA 121
EQ R++ +QQ LL+++R+ ++ +YYA
Sbjct 319 EQIRIEREQQKKLLKKERKTLRDKVKDCKYYA 350
> SPBC1778.01c
Length=354
Score = 35.4 bits (80), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 37/74 (50%), Gaps = 2/74 (2%)
Query 3 GDVGPFYSYWHSFTSVRDFAEVDQWSPKDMAEASRAERRLMERDNGRKRKEAKKEFVDTV 62
+V FY++W++F S R F +D+ P D SR +R E+ N +R++ K +
Sbjct 126 AEVDNFYNFWYNFDSWRSFEYLDKDIPDD--GESRDNKRFQEKKNRSERQKNKARDNARL 183
Query 63 RRLAQHIKKKDPRV 76
R L DPR+
Sbjct 184 RNLVDTALASDPRI 197
> YER008c
Length=1336
Score = 31.6 bits (70), Expect = 0.34, Method: Composition-based stats.
Identities = 24/90 (26%), Positives = 48/90 (53%), Gaps = 12/90 (13%)
Query 51 RKEAKKEFVDTVRRLAQHIKKKD-----PRVLARQAALAQQRLDEQRLKE-------EEQ 98
+KE + + ++ RR+ Q +K+ R L + Q L+ ++ E EE+
Sbjct 354 QKENEMKRLEEERRIKQEERKRQMELEHQRQLEEEERKRQMELEAKKQMELKRQRQFEEE 413
Query 99 QRVQEQQQLLRQQRQQQQEEYYAALKAERR 128
QR++++++LL QR+Q+++E LK E +
Sbjct 414 QRLKKERELLEIQRKQREQETAERLKKEEQ 443
> ECU01g0370
Length=295
Score = 30.0 bits (66), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 34/69 (49%), Gaps = 5/69 (7%)
Query 8 FYSYWHSFTSVRDFAEVDQWSPKDMAEASRAERRLMERDNGRKRKEAKKEFVDTVRRLAQ 67
FY +W +F S R F V++ M E R++ R+ K K + ++RL Q
Sbjct 162 FYEFWSNFRSWRTFEPVEELY--GMEEHDRSQYSAKNRE---KLASLKNQDALRIKRLVQ 216
Query 68 HIKKKDPRV 76
KK+DPR+
Sbjct 217 IAKKRDPRI 225
> SPAC6B12.08
Length=380
Score = 29.6 bits (65), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 22/76 (28%), Positives = 34/76 (44%), Gaps = 6/76 (7%)
Query 2 WGDVGPFYSYWHSFTSVRDFAEVDQWSP--KDMAEASRAERRLMERDNGRKRKEAKKEFV 59
W P Y W F++ + F +W + E+ A RRLM+R N R+ + + +
Sbjct 131 WTYAKPIYQKWLRFSTKKSF----EWEALYNEEEESDAATRRLMKRQNQRQIQYCIQRYN 186
Query 60 DTVRRLAQHIKKKDPR 75
+ VR L DPR
Sbjct 187 ELVRDLIGKACDLDPR 202
> Hs22062520
Length=1083
Score = 27.7 bits (60), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 37/71 (52%), Gaps = 7/71 (9%)
Query 53 EAKKEFVDTVRRLAQHIKKKDPRVLARQAALAQQRLDEQRLKEEEQQRVQEQQQLLRQQR 112
E E +V RL +++K +L Q ALAQQR R RV+E ++ L +Q
Sbjct 564 ELGSEVSTSVMRLKLEVEEKKQAMLLLQRALAQQRDLTAR-------RVKETEKALSRQL 616
Query 113 QQQQEEYYAAL 123
Q+Q+E Y A +
Sbjct 617 QRQREHYEATI 627
Lambda K H
0.317 0.128 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1213511838
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40