bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1558_orf1
Length=140
Score E
Sequences producing significant alignments: (Bits) Value
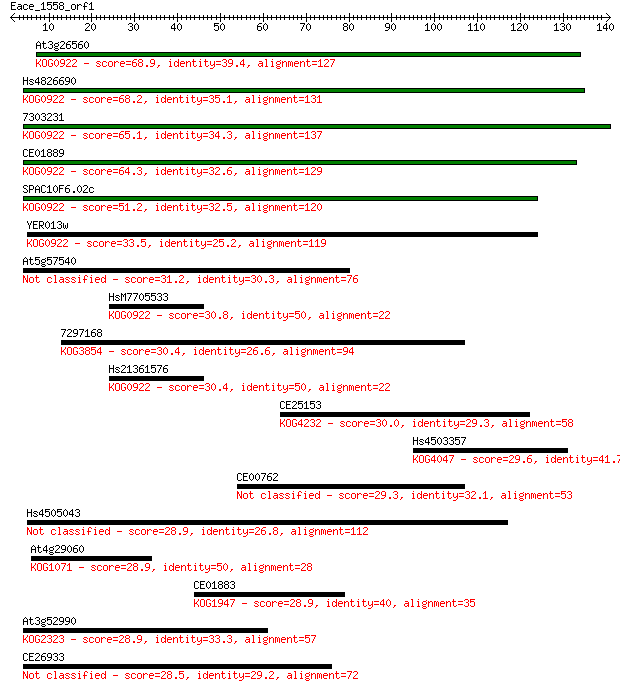
At3g26560 68.9 3e-12
Hs4826690 68.2 4e-12
7303231 65.1 4e-11
CE01889 64.3 7e-11
SPAC10F6.02c 51.2 5e-07
YER013w 33.5 0.15
At5g57540 31.2 0.65
HsM7705533 30.8 0.90
7297168 30.4 1.1
Hs21361576 30.4 1.1
CE25153 30.0 1.3
Hs4503357 29.6 1.9
CE00762 29.3 2.5
Hs4505043 28.9 3.0
At4g29060 28.9 3.2
CE01883 28.9 3.3
At3g52990 28.9 3.6
CE26933 28.5 4.7
> At3g26560
Length=1168
Score = 68.9 bits (167), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 50/129 (38%), Positives = 70/129 (54%), Gaps = 12/129 (9%)
Query 7 VKKGQLVKVKVHAIAGSRLSLTMREVDQATGRDLKPRNLQSGNAAGSRAALFESLAEEHA 66
VK+ V VKV +I+ + SL+MR+VDQ TGRDL P S SR+ S +
Sbjct 260 VKRDMEVYVKVISISSDKYSLSMRDVDQNTGRDLIPLRKPSDEDDSSRSN--PSYRTKDG 317
Query 67 QALEKKGLGRITGVKINIESTEEETAYARKRKL--MSDYDKWEAQQLQRSGLVSSKENPY 124
Q + K G I+G++I EE +R L MS ++WEA+QL SG++ E P
Sbjct 318 Q-VTKTG---ISGIRI----VEENDVAPSRRPLKKMSSPERWEAKQLIASGVLRVDEFPM 369
Query 125 YDEEAGGLL 133
YDE+ G+L
Sbjct 370 YDEDGDGML 378
> Hs4826690
Length=1220
Score = 68.2 bits (165), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 46/132 (34%), Positives = 70/132 (53%), Gaps = 15/132 (11%)
Query 4 ADAVKKGQLVKVKVHAIAGSRLSLTMREVDQATGRDLKPRNLQSGNAAGSRAALFESLAE 63
AD V KGQ VKVKV + G++ SL+M++VDQ TG DL P + +L
Sbjct 310 ADVVSKGQRVKVKVLSFTGTKTSLSMKDVDQETGEDLNPNRRR-------------NLVG 356
Query 64 EHAQALEKKGLGRITGVKINIESTEEETAYARKR-KLMSDYDKWEAQQLQRSGLVSSKEN 122
E + + R T + + E+ + RKR +SD +KWE +Q+ + ++S +E
Sbjct 357 ETNEETSMRNPDRPTHLSLVSAPEVEDDSLERKRLTRISDPEKWEIKQMIAANVLSKEEF 416
Query 123 PYYDEEAGGLLP 134
P +DEE G+LP
Sbjct 417 PDFDEET-GILP 427
> 7303231
Length=1242
Score = 65.1 bits (157), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 47/138 (34%), Positives = 71/138 (51%), Gaps = 13/138 (9%)
Query 4 ADAVKKGQLVKVKVHAIAGSRLSLTMREVDQATGRDLKPRNLQSGNAAGSRAALFESLAE 63
+ V + Q VKVKV +I G ++SL+M+EVDQ +G+DL P + + R +
Sbjct 330 TEVVTRNQTVKVKVMSITGQKVSLSMKEVDQDSGKDLNPLSHAPEDDESLRDRNPDGPFS 389
Query 64 EHAQALEKKGLGRITGVKINIESTEEETAYARKRKL-MSDYDKWEAQQLQRSGLVSSKEN 122
L +G G +E E E +RKR +S ++WE +Q+ SG++ E
Sbjct 390 SSTSMLNLQGNG--------MEGDEHE---SRKRVTRISSPERWEIKQMISSGVLDRSEM 438
Query 123 PYYDEEAGGLLPSQEAEE 140
P +DEE GLLP E +E
Sbjct 439 PDFDEET-GLLPKDEDDE 455
> CE01889
Length=1200
Score = 64.3 bits (155), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 42/130 (32%), Positives = 69/130 (53%), Gaps = 11/130 (8%)
Query 4 ADAVKKGQLVKVKVHAIAGSRLSLTMREVDQATGRDLKPRNLQ-SGNAAGSRAALFESLA 62
AD +K+G+ VKVKV+ I ++SL+M+EVDQ +G DL PR + +A G R +
Sbjct 278 ADVLKRGENVKVKVNKIENGKISLSMKEVDQNSGEDLNPRETDLNPDAIGVRPRTPPAST 337
Query 63 EEHAQALEKKGLGRITGVKINIESTEEETAYARKRKLMSDYDKWEAQQLQRSGLVSSKEN 122
E G+G+ I + R +S ++WE +Q+Q +G++++ +
Sbjct 338 SSWMNP-EASGVGQGPSTSI---------GGGKARVRISTPERWELRQMQGAGVLTATDM 387
Query 123 PYYDEEAGGL 132
P +DEE G L
Sbjct 388 PDFDEEMGVL 397
> SPAC10F6.02c
Length=1168
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 39/122 (31%), Positives = 62/122 (50%), Gaps = 18/122 (14%)
Query 4 ADAVKKGQLVKVKVHAI--AGSRLSLTMREVDQATGRDLKPRNLQSGNAAGSRAALFESL 61
++AV GQ V VKV I + R+SL+M+EV+Q TG DL P + GS A L
Sbjct 252 SEAVSYGQPVFVKVIRIDESAKRISLSMKEVNQVTGEDLNPDQVSRSTKKGSGANAI-PL 310
Query 62 AEEHAQALEKKGLGRITGVKINIESTEEETAYARKRKLMSDYDKWEAQQLQRSGLVSSKE 121
+ ++++ +G + + ET + RK ++ + WE QQL SG +S+ +
Sbjct 311 SAQNSE------IGHVNPL---------ETFTSNGRKRLTSPEIWELQQLAASGAISATD 355
Query 122 NP 123
P
Sbjct 356 IP 357
> YER013w
Length=1145
Score = 33.5 bits (75), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 30/120 (25%), Positives = 55/120 (45%), Gaps = 30/120 (25%)
Query 5 DAVKKGQLVKVKVHAIAGS-RLSLTMREVDQATGRDLKPRNLQSGNAAGSRAALFESLAE 63
D V++GQ + V+V I + ++SL+M+ +DQ +G +++ RN +S
Sbjct 224 DVVRQGQHIFVEVIKIQNNGKISLSMKNIDQHSG-EIRKRNTES---------------- 266
Query 64 EHAQALEKKGLGRITGVKINIESTEEETAYARKRKLMSDYDKWEAQQLQRSGLVSSKENP 123
+E +G + N T KR+ ++ ++WE +QL SG S + P
Sbjct 267 -----VEDRG-------RSNDAHTSRNMKNKIKRRALTSPERWEIRQLIASGAASIDDYP 314
> At5g57540
Length=284
Score = 31.2 bits (69), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 41/77 (53%), Gaps = 5/77 (6%)
Query 4 ADAVKKGQLVKVKVHAIAGSRLSLTMREVDQATGR-DLKPRNLQSGNAAGSRAALFESLA 62
A+ V+ GQL+ + I+GS + + G+ D+K + L +GN+AG+ A + S
Sbjct 39 ANIVESGQLLTCTLDKISGSGFQ---SKKEYLFGKIDMKMK-LVAGNSAGTVTAYYLSSK 94
Query 63 EEHAQALEKKGLGRITG 79
E ++ + LG +TG
Sbjct 95 GETWDEIDFEFLGNVTG 111
> HsM7705533
Length=212
Score = 30.8 bits (68), Expect = 0.90, Method: Compositional matrix adjust.
Identities = 11/22 (50%), Positives = 18/22 (81%), Gaps = 0/22 (0%)
Query 24 RLSLTMREVDQATGRDLKPRNL 45
++SL+M+ V+Q TG+DL P N+
Sbjct 53 KVSLSMKVVNQGTGKDLDPNNV 74
> 7297168
Length=449
Score = 30.4 bits (67), Expect = 1.1, Method: Composition-based stats.
Identities = 25/98 (25%), Positives = 43/98 (43%), Gaps = 8/98 (8%)
Query 13 VKVKVHAIAG---SRLSLTMREVDQATGRDLKPRNLQSGN-AAGSRAALFESLAEEHAQA 68
+K K+H G SR L + A D+KP ++++ N S A E Q
Sbjct 70 LKTKMHKFLGLIPSRRKL----YNPAVVYDMKPEDIENINCVVNSSHASAEPFTPSAEQP 125
Query 69 LEKKGLGRITGVKINIESTEEETAYARKRKLMSDYDKW 106
+ K + T K+N+ + +E T + K +D++ W
Sbjct 126 IRPKTMMTSTWSKLNLSNIDEPTPEESQAKSKNDFELW 163
> Hs21361576
Length=241
Score = 30.4 bits (67), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 11/22 (50%), Positives = 18/22 (81%), Gaps = 0/22 (0%)
Query 24 RLSLTMREVDQATGRDLKPRNL 45
++SL+M+ V+Q TG+DL P N+
Sbjct 82 KVSLSMKVVNQGTGKDLDPNNV 103
> CE25153
Length=443
Score = 30.0 bits (66), Expect = 1.3, Method: Composition-based stats.
Identities = 17/58 (29%), Positives = 34/58 (58%), Gaps = 3/58 (5%)
Query 64 EHAQALEKKGLGRITGVKINIESTEEETAYARKRKLMSDYDKWEAQQLQRSGLVSSKE 121
EH + LEK+ R+ V IN+ + + + A+++K++ ++K Q+L GL+ + E
Sbjct 69 EHDEFLEKQLEKRLDKVDINVSAY--DVSVAQEKKMVESFEKLR-QKLHDDGLMKANE 123
> Hs4503357
Length=481
Score = 29.6 bits (65), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 95 RKRKLMSDYDKWEAQQLQRSGLVSSKENPYYDEEAG 130
RK+ L D + QQL ++ L KE+P YDE G
Sbjct 332 RKKPLYWDLYEHAQQQLLKAKLTDPKEDPIYDEPEG 367
> CE00762
Length=934
Score = 29.3 bits (64), Expect = 2.5, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 32/53 (60%), Gaps = 3/53 (5%)
Query 54 RAALFESLAEEHAQALEKKGLGRITGVKINIESTEEETAYARKRKLMSDYDKW 106
+ AL +++ E +A + KK + + G IN+ +++ E Y R+RKL S + +W
Sbjct 675 KNALLDAVREINATNV-KKAVNQ--GAYINVFNSQFEIIYYRRRKLKSPFTEW 724
> Hs4505043
Length=711
Score = 28.9 bits (63), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 30/126 (23%), Positives = 54/126 (42%), Gaps = 15/126 (11%)
Query 5 DAVKKGQLVKVKVHAIAGSRLS-------LTMREVDQATGRDLKPRNLQSGNAAGSRAAL 57
D K L +V HA+ ++ +R+ + G+D P+ G+ +G + L
Sbjct 260 DKFKDCHLARVPSHAVVARSVNGKEDAIWNLLRQAQEKFGKDKSPKFQLFGSPSGQKDLL 319
Query 58 FESLAEEHAQALEKK------GLGRITGVKINIESTEEETAYARKRKLMSDYDKWEAQQL 111
F+ A ++ + G G T ++ N+ +EEE A R R + + E ++
Sbjct 320 FKDSAIGFSRVPPRIDSGLYLGSGYFTAIQ-NLRKSEEEVAARRARVVWCAVGEQELRKC 378
Query 112 -QRSGL 116
Q SGL
Sbjct 379 NQWSGL 384
> At4g29060
Length=953
Score = 28.9 bits (63), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 14/28 (50%), Positives = 21/28 (75%), Gaps = 0/28 (0%)
Query 6 AVKKGQLVKVKVHAIAGSRLSLTMREVD 33
+++ GQ VKV+V IA R++LTM+E D
Sbjct 294 SLQAGQEVKVRVLRIARGRVTLTMKEED 321
> CE01883
Length=699
Score = 28.9 bits (63), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 44 NLQSGNAAGSRAALFESLAEEHAQALEKKGLGRIT 78
NL S +A S+ ++F++L H Q LE +GL +T
Sbjct 10 NLGSAGSAKSQQSMFKALRRLHGQNLESRGLHDMT 44
> At3g52990
Length=514
Score = 28.9 bits (63), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 26/57 (45%), Gaps = 0/57 (0%)
Query 4 ADAVKKGQLVKVKVHAIAGSRLSLTMREVDQATGRDLKPRNLQSGNAAGSRAALFES 60
A AVKKG + V + GS + EVD+ G D+ + + AGS L S
Sbjct 126 AKAVKKGDTIFVGQYLFTGSETTSVWLEVDEVKGDDVICLSRNAATLAGSLFTLHSS 182
> CE26933
Length=211
Score = 28.5 bits (62), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 36/74 (48%), Gaps = 5/74 (6%)
Query 4 ADAVKKGQLVKVKVHAIAGSRLSLTMREVDQAT--GRDLKPRNLQSGNAAGSRAALFESL 61
+ +VKKG++V K T + +++A G L P ++G S++ + L
Sbjct 99 SPSVKKGEIVAPKATVTPAVN---TRKPINEAISEGIILDPTYFKNGGNLDSKSPKNQKL 155
Query 62 AEEHAQALEKKGLG 75
AEE Q ++ GLG
Sbjct 156 AEELQQFIDNPGLG 169
Lambda K H
0.308 0.125 0.333
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1571531578
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40