bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1438_orf2
Length=88
Score E
Sequences producing significant alignments: (Bits) Value
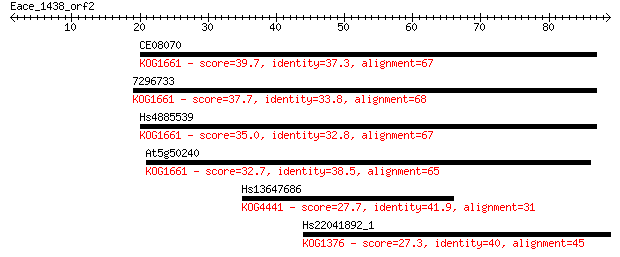
CE08070 39.7 0.001
7296733 37.7 0.005
Hs4885539 35.0 0.033
At5g50240 32.7 0.19
Hs13647686 27.7 5.3
Hs22041892_1 27.3 7.7
> CE08070
Length=225
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 31/67 (46%), Gaps = 18/67 (26%)
Query 20 ALLQQLAAGGRMVIPVECNRGFISLEGLGDREVGGCDGGQALVSVEKDKMGNVSVKYITS 79
AL QLA GGRM+IPVE DG Q + ++K G + K +
Sbjct 168 ALTDQLAEGGRMMIPVEQ-----------------VDGNQVFMQIDKIN-GKIEQKIVEH 209
Query 80 VIYVPLV 86
VIYVPL
Sbjct 210 VIYVPLT 216
> 7296733
Length=226
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 34/69 (49%), Gaps = 19/69 (27%)
Query 19 SALLQQLAAGGRMVIPVECNRGFISLEGLGDREVGGCDGG-QALVSVEKDKMGNVSVKYI 77
+ L+ QLA+GGR+++PV G DGG Q + +KD G V + +
Sbjct 172 TELINQLASGGRLIVPV------------------GPDGGSQYMQQYDKDANGKVEMTRL 213
Query 78 TSVIYVPLV 86
V+YVPL
Sbjct 214 MGVMYVPLT 222
> Hs4885539
Length=227
Score = 35.0 bits (79), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 32/67 (47%), Gaps = 17/67 (25%)
Query 20 ALLQQLAAGGRMVIPVECNRGFISLEGLGDREVGGCDGGQALVSVEKDKMGNVSVKYITS 79
AL+ QL GGR+++PV G G Q L +K + G++ +K +
Sbjct 168 ALIDQLKPGGRLILPV-----------------GPAGGNQMLEQYDKLQDGSIKMKPLMG 210
Query 80 VIYVPLV 86
VIYVPL
Sbjct 211 VIYVPLT 217
> At5g50240
Length=269
Score = 32.7 bits (73), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 31/65 (47%), Gaps = 19/65 (29%)
Query 21 LLQQLAAGGRMVIPVECNRGFISLEGLGDREVGGCDGGQALVSVEKDKMGNVSVKYITSV 80
LL QL GGRMVIP LG Q L ++K++ G++ V TSV
Sbjct 214 LLDQLKPGGRMVIP------------LGTY-------FQELKVIDKNEDGSIKVHTETSV 254
Query 81 IYVPL 85
YVPL
Sbjct 255 RYVPL 259
> Hs13647686
Length=268
Score = 27.7 bits (60), Expect = 5.3, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 20/33 (60%), Gaps = 2/33 (6%)
Query 35 VECNRGFISLEGLGDR--EVGGCDGGQALVSVE 65
+ C R +++ L D+ +GGCDG +L SVE
Sbjct 216 ITCKRCYVASVSLHDQIYVIGGCDGRSSLSSVE 248
> Hs22041892_1
Length=193
Score = 27.3 bits (59), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 25/47 (53%), Gaps = 5/47 (10%)
Query 44 LEGLGDREVGGCDGGQALVSVEKDKMGNVS--VKYITSVIYVPLVRT 88
+GLGD E+ GC G V+V +D G + V +IT + L RT
Sbjct 115 WQGLGDNELDGCSGE---VNVSQDFCGKYTDVVNHITQRLDYVLCRT 158
Lambda K H
0.320 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1184307974
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40