bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1415_orf1
Length=160
Score E
Sequences producing significant alignments: (Bits) Value
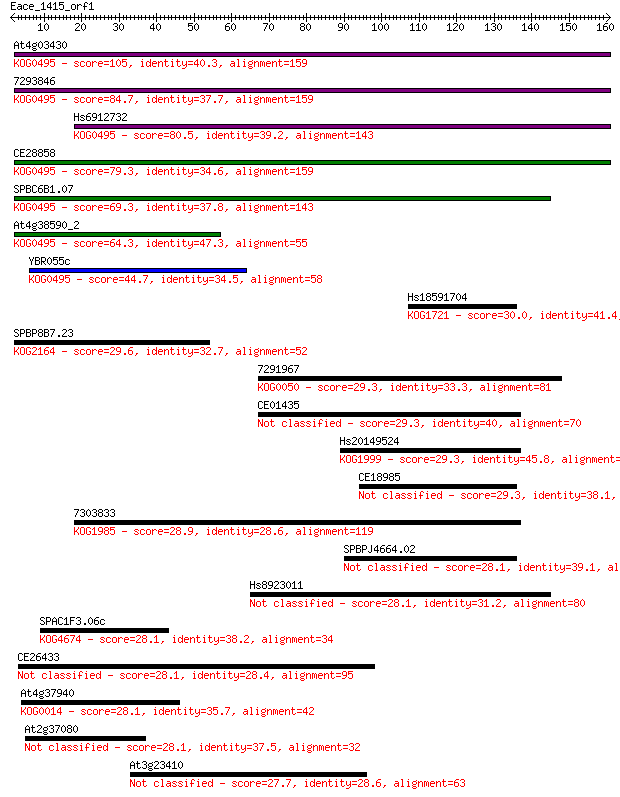
At4g03430 105 3e-23
7293846 84.7 6e-17
Hs6912732 80.5 1e-15
CE28858 79.3 3e-15
SPBC6B1.07 69.3 3e-12
At4g38590_2 64.3 1e-10
YBR055c 44.7 8e-05
Hs18591704 30.0 2.2
SPBP8B7.23 29.6 2.7
7291967 29.3 3.4
CE01435 29.3 3.6
Hs20149524 29.3 3.7
CE18985 29.3 3.9
7303833 28.9 4.8
SPBPJ4664.02 28.1 6.7
Hs8923011 28.1 7.0
SPAC1F3.06c 28.1 7.0
CE26433 28.1 7.0
At4g37940 28.1 7.5
At2g37080 28.1 8.3
At3g23410 27.7 10.0
> At4g03430
Length=1029
Score = 105 bits (263), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 64/159 (40%), Positives = 88/159 (55%), Gaps = 29/159 (18%)
Query 2 RRRSRREARLKEEIAKLRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDYTLKRKQKK 61
RR+ RREA+LKEEI K RA P I +QFADLK L +++ +EW+SIP+IGDY+L+ K+KK
Sbjct 212 RRKDRREAKLKEEIEKYRASNPKITEQFADLKRKLHTLSADEWDSIPEIGDYSLRNKKKK 271
Query 62 QQMLTPASDRGILEARNRESFSNSIASAGSATPIGFGMATPLMAGGLSTPLGLQTPLGIQ 121
+ P D + +A+ + ++ A AGG TP G Q
Sbjct 272 FESFVPIPDTLLEKAKKEKELVMALDPKSRA------------AGGSETPWG-------Q 312
Query 122 TPLGLRTPMGSSTPSLNDLGEARGTVLSVKLDKVMDSIS 160
TP+ L +GE RGTVLS+KLD + DS+S
Sbjct 313 TPV----------TDLTAVGEGRGTVLSLKLDNLSDSVS 341
> 7293846
Length=913
Score = 84.7 bits (208), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 60/161 (37%), Positives = 89/161 (55%), Gaps = 28/161 (17%)
Query 2 RRRSRREARLKEEIAKLRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDYTLKRKQK- 60
+R+ R+ RL+E++ + R E+P I QQF+DLK L SVT EEW +IP++GD + RKQ+
Sbjct 119 KRKEYRDRRLREDLERYRQERPKIQQQFSDLKRSLASVTSEEWSTIPEVGD-SRNRKQRN 177
Query 61 -KQQMLTPASDRGILEARNRESFSNSIASAGSATPIGFGMATPLMAGGLSTPLGLQTPLG 119
+ + TP D I ES S S+G A+ M G++TP G+ TP G
Sbjct 178 PRAEKFTPLPDSLISRNLGGESSSTLDPSSGLAS----------MVPGVATP-GMLTPTG 226
Query 120 IQTPLGLRTPMGSSTPSLNDLGEARGTVLSVKLDKVMDSIS 160
L LR +G+AR T+++VKL +V DS++
Sbjct 227 ---DLDLRK-----------IGQARNTLMNVKLSQVSDSVT 253
> Hs6912732
Length=941
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 56/145 (38%), Positives = 82/145 (56%), Gaps = 14/145 (9%)
Query 18 LRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDYTLKRKQK-KQQMLTPASDRGILEA 76
R E+P I QQF+DLK L VT+EEW SIP++GD KR++ + + LTP D A
Sbjct 131 YRMERPKIQQQFSDLKRKLAEVTEEEWLSIPEVGDARNKRQRNPRYEKLTPVPDSFF--A 188
Query 77 RNRESFSNSIASAGSATPIGFGMATPLMAGGLSTPL-GLQTPLGIQTPLGLRTPMGSSTP 135
++ ++ N + T G G+ TP GGL+TP G TP GL TP G+
Sbjct 189 KHLQTGENHTSVDPRQTQFG-GLNTP-YPGGLNTPYRGGMTP-------GLMTP-GTGEL 238
Query 136 SLNDLGEARGTVLSVKLDKVMDSIS 160
+ +G+AR T++ ++L +V DS+S
Sbjct 239 DMRKIGQARNTLMDMRLSQVSDSVS 263
> CE28858
Length=968
Score = 79.3 bits (194), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 55/169 (32%), Positives = 83/169 (49%), Gaps = 17/169 (10%)
Query 2 RRRSRREARLKEEIAKLRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDYTLKRKQK- 60
R + RRE + KE + ++P I Q F DLK L VT++EW++IP++GD K K+
Sbjct 128 RHKERREKKYKELVELFHKDRPKIQQGFQDLKRQLAEVTEDEWQAIPEVGDMRNKAKRNA 187
Query 61 KQQMLTPASDRGILEARNRESFSNSIASAGSATPIGFGMATPLMAGGLSTPLGLQTPLGI 120
+ + TP D I N S+SI S G G+ TP +G +ST G
Sbjct 188 RAEKFTPVPDSIIAMNMNYGQMSHSIDS-------GNGLTTPFASGFMSTLGGGGGAAAA 240
Query 121 QTPLGLRTP-MGSSTPS--------LNDLGEARGTVLSVKLDKVMDSIS 160
G+ TP S PS L +G+AR ++ ++L +V DS++
Sbjct 241 GAKSGIMTPGWKSDIPSSGTSTDLDLVKIGQARNKIMDMQLTQVSDSVT 289
> SPBC6B1.07
Length=906
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 54/160 (33%), Positives = 82/160 (51%), Gaps = 18/160 (11%)
Query 2 RRRSRREARLKEEIAKLRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDYTLKR--KQ 59
RR+S+RE + + + K E P + QFADLK GL+++T E+W +IP+ GD T K+ KQ
Sbjct 103 RRKSQREKQEQLQKEKYEKENPKVSSQFADLKRGLSTLTDEDWNNIPEPGDLTRKKRTKQ 162
Query 60 KKQQMLTPASDRGILEARNRESFSNSIA---SAGSATPIGFGMATPLMAGGLS--TPLGL 114
+++ SD + ARN ++ A AG+ TP G T + G + LG+
Sbjct 163 PRRERFYATSDFVLASARNENQAISNFAVDTQAGTETPDMNGTKTNFVEIGAARDKVLGI 222
Query 115 Q---------TPLGIQTPLGLRTPMGSSTP-SLNDLGEAR 144
+ +P I P G T + S P + NDLG+ R
Sbjct 223 KLAQASSNLTSPSTID-PKGYLTSLNSMVPKNANDLGDIR 261
> At4g38590_2
Length=160
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 26/55 (47%), Positives = 40/55 (72%), Gaps = 0/55 (0%)
Query 2 RRRSRREARLKEEIAKLRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDYTLK 56
RR+ RREA+LK+EI RA P + QF DL L +++++EW+SIP+IG+Y+ +
Sbjct 104 RRKDRREAKLKQEIENYRASNPKVSGQFVDLTRKLHTLSEDEWDSIPEIGNYSHR 158
> YBR055c
Length=899
Score = 44.7 bits (104), Expect = 8e-05, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 33/58 (56%), Gaps = 0/58 (0%)
Query 6 RREARLKEEIAKLRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDYTLKRKQKKQQ 63
+++ R E+ + + +QFADLK L +VT+ EW IPD D+T + K+ + Q
Sbjct 81 KKKKRANEKDDDNSVDSSNVKRQFADLKESLAAVTESEWMDIPDATDFTRRNKRNRIQ 138
> Hs18591704
Length=851
Score = 30.0 bits (66), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 12/29 (41%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 107 GLSTPLGLQTPLGIQTPLGLRTPMGSSTP 135
G+S P+G+ P+ I P+G+ P+G S P
Sbjct 146 GVSKPIGVSKPVTIGKPVGVSKPIGISKP 174
> SPBP8B7.23
Length=673
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 31/56 (55%), Gaps = 4/56 (7%)
Query 2 RRRSRREARLKEEIAKLRAEKPTIHQQFADLKMGL----TSVTQEEWESIPDIGDY 53
RR+ R+ ++EE + RAE+ T Q +++L M T V ++ E+ P +G +
Sbjct 539 RRKQRKAQEMREERYRQRAERDTEEQIYSELNMQRPVPKTVVHEDPAEAFPTLGSH 594
> 7291967
Length=791
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 27/98 (27%), Positives = 37/98 (37%), Gaps = 23/98 (23%)
Query 67 PASDRGILEARNRESFSNSIASAGSATPIGFGMATPLMAGGLS----------------- 109
P +DR + EA+N +A + TP+ G+ TPL S
Sbjct 327 PYTDRIMQEAQN------MMALTHTETPLKGGLNTPLHESDFSGVLPKAASIATPNTVIA 380
Query 110 TPLGLQTPLGIQTPLGLRTPMGSSTPSLNDLGEARGTV 147
TP Q G TP G +TP + + G A G V
Sbjct 381 TPFRTQREGGAATPGGFQTPSSGALVPVKGAGGATGVV 418
> CE01435
Length=213
Score = 29.3 bits (64), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 28/76 (36%), Positives = 38/76 (50%), Gaps = 7/76 (9%)
Query 67 PASDRGILEARNRESFSNSIASAGSATPI-GF----GMATPLMAGGLS-TPLGLQTPLGI 120
P S + E R+R S S SA+P+ GF G T + G + P G+Q PL
Sbjct 11 PQSSISLDEKRHRNR-SLGCISRYSASPLPGFSAFLGTITRVSTGQTTFHPYGVQEPLSP 69
Query 121 QTPLGLRTPMGSSTPS 136
Q+ +G+ T M STPS
Sbjct 70 QSCVGVFTSMYCSTPS 85
> Hs20149524
Length=1087
Score = 29.3 bits (64), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 22/52 (42%), Positives = 28/52 (53%), Gaps = 5/52 (9%)
Query 89 AGSATPIGFGMATPLMAGGLSTPLGLQTPL--GIQTPL--GLRTPMGSSTPS 136
+GS TP+ +G TPL G + G QTPL G +TP G P +TPS
Sbjct 780 SGSRTPM-YGSQTPLQDGSRTPHYGSQTPLHDGSRTPAQSGAWDPNNPNTPS 830
> CE18985
Length=627
Score = 29.3 bits (64), Expect = 3.9, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 25/48 (52%), Gaps = 6/48 (12%)
Query 94 PIGFG------MATPLMAGGLSTPLGLQTPLGIQTPLGLRTPMGSSTP 135
PI G +A L+A G+ PL + TP + TP + TP+ ++ P
Sbjct 533 PISLGNINFVALAQQLIASGIKLPLPIVTPPVVSTPAPVITPIPAALP 580
> 7303833
Length=1184
Score = 28.9 bits (63), Expect = 4.8, Method: Composition-based stats.
Identities = 34/138 (24%), Positives = 51/138 (36%), Gaps = 25/138 (18%)
Query 18 LRAEKPTIHQQFADLKMGLTSVTQEEWESIPDIGDYTLKRKQKKQQ---MLTPASDRGIL 74
LR HQ A+L L ++ + + G L + Q +QQ +T RG+
Sbjct 162 LRPSPQPTHQSLANLTSNLENLNLNTTRA--NGGTSILPQPQTQQQYAPTVTQKDGRGVA 219
Query 75 EARNRESFSNSIASAGSATPIGFGMATPLMAGGLSTPLGLQTP----------------L 118
A+ N S +A+ G+A L+ ++TP Q P
Sbjct 220 SAQTGPPQFNDANSLLAAS----GVAAALVPNAMATPFSGQKPPNLAPNPAKMFGSASMA 275
Query 119 GIQTPLGLRTPMGSSTPS 136
G TP G R P G P+
Sbjct 276 GRPTPAGQRIPYGVPPPT 293
> SPBPJ4664.02
Length=3971
Score = 28.1 bits (61), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 27/53 (50%), Gaps = 7/53 (13%)
Query 90 GSATPI----GFGMATPLMAGGL---STPLGLQTPLGIQTPLGLRTPMGSSTP 135
S+TPI G +TP+ + + STP+ T L TP+ T + +STP
Sbjct 931 NSSTPITSSTGLNTSTPITSSSVLNSSTPITSSTVLNSSTPITSSTALNTSTP 983
> Hs8923011
Length=294
Score = 28.1 bits (61), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 34/80 (42%), Gaps = 10/80 (12%)
Query 65 LTPASDRGILEARNRESFSNSIASAGSATPIGFGMATPLMAGGLSTPLGLQTPLGIQTPL 124
P +R +L+AR R +ATP G G TP AG ++ +G+ P
Sbjct 120 FNPRVERTLLDARRR-------LEDQAATPTGLGSLTPSAAGSTASLVGVGLPPPTPRSS 172
Query 125 GLRTPMGSSTPSLNDLGEAR 144
GL TP+ PS L R
Sbjct 173 GLSTPV---PPSAGHLAHVR 189
> SPAC1F3.06c
Length=1957
Score = 28.1 bits (61), Expect = 7.0, Method: Composition-based stats.
Identities = 13/34 (38%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 9 ARLKEEIAKLRAEKPTIHQQFADLKMGLTSVTQE 42
++L+EE+ +L+ TIH Q D + L+S QE
Sbjct 367 SQLEEEMVELKESNRTIHSQLTDAESKLSSFEQE 400
> CE26433
Length=613
Score = 28.1 bits (61), Expect = 7.0, Method: Composition-based stats.
Identities = 27/107 (25%), Positives = 47/107 (43%), Gaps = 15/107 (14%)
Query 3 RRSRREARLKEEIAKLRAEKPTIHQQF-ADLKMGLTSVTQEEWESIPDIGDYTLKRKQKK 61
RR RL+ +L + H++ ++ + V +EE ++ P GD + K K+
Sbjct 304 RRITVNRRLRS--GELEKNEQKEHKELRVKMREEIDKVIEEECKNKPK-GDISAGEKPKQ 360
Query 62 QQML-----------TPASDRGILEARNRESFSNSIASAGSATPIGF 97
++L T A + G+ R+ F N ++GS PIGF
Sbjct 361 ARLLNSSAEFFDRFETKAEEAGLSRKEFRDPFFNIKLNSGSHVPIGF 407
> At4g37940
Length=228
Score = 28.1 bits (61), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 23/42 (54%), Gaps = 0/42 (0%)
Query 4 RSRREARLKEEIAKLRAEKPTIHQQFADLKMGLTSVTQEEWE 45
R R+E L +EI +L ++ IHQ+ DL + + QE E
Sbjct 138 RMRKEQLLTQEIQELSQKRNLIHQENLDLSRKVQRIHQENVE 179
> At2g37080
Length=583
Score = 28.1 bits (61), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 12/32 (37%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 5 SRREARLKEEIAKLRAEKPTIHQQFADLKMGL 36
++REA L EE+ K +AE+ ++H++ D + L
Sbjct 339 AQREAELGEELKKTKAERDSLHERLMDKEAKL 370
> At3g23410
Length=746
Score = 27.7 bits (60), Expect = 10.0, Method: Compositional matrix adjust.
Identities = 18/63 (28%), Positives = 30/63 (47%), Gaps = 4/63 (6%)
Query 33 KMGLTSVTQEEWESIPDIGDYTLKRKQKKQQMLTPASDRGILEARNRESFSNSIASAGSA 92
K GL +T E ES+ + L Q + L+ D RN+E+ + +++GS
Sbjct 9 KFGLPDITVAEMESLASFCEAVLPSVQPPPEELSGEGDN----HRNKEALRSFYSTSGSK 64
Query 93 TPI 95
TP+
Sbjct 65 TPV 67
Lambda K H
0.312 0.130 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2172509160
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40