bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1379_orf1
Length=133
Score E
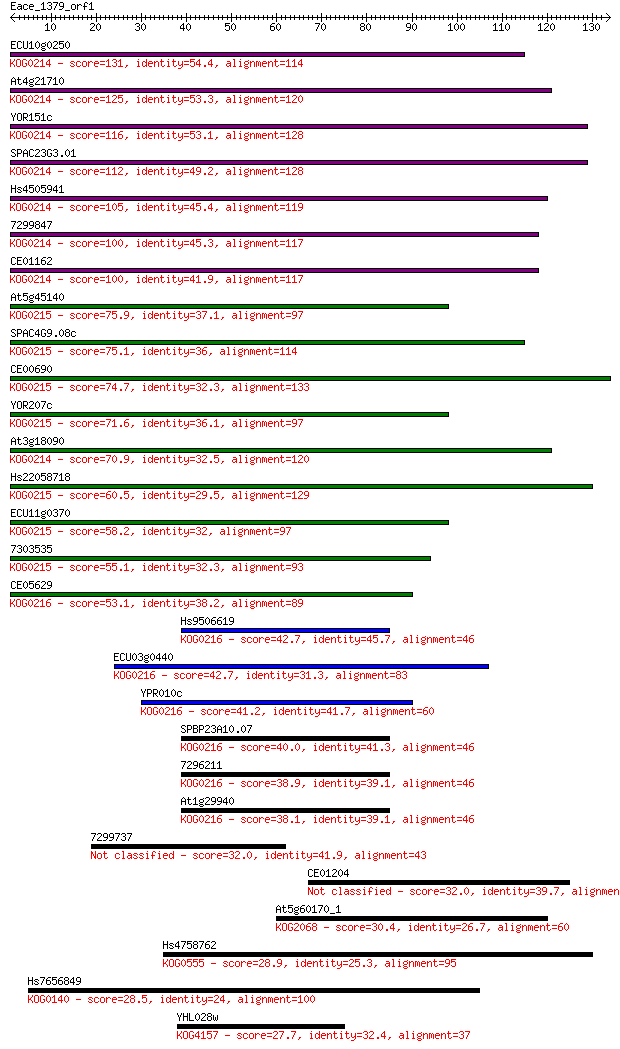
Sequences producing significant alignments: (Bits) Value
ECU10g0250 131 4e-31
At4g21710 125 3e-29
YOR151c 116 1e-26
SPAC23G3.01 112 1e-25
Hs4505941 105 2e-23
7299847 100 8e-22
CE01162 100 9e-22
At5g45140 75.9 2e-14
SPAC4G9.08c 75.1 3e-14
CE00690 74.7 4e-14
YOR207c 71.6 3e-13
At3g18090 70.9 6e-13
Hs22058718 60.5 8e-10
ECU11g0370 58.2 4e-09
7303535 55.1 3e-08
CE05629 53.1 1e-07
Hs9506619 42.7 2e-04
ECU03g0440 42.7 2e-04
YPR010c 41.2 6e-04
SPBP23A10.07 40.0 0.001
7296211 38.9 0.003
At1g29940 38.1 0.004
7299737 32.0 0.32
CE01204 32.0 0.36
At5g60170_1 30.4 1.1
Hs4758762 28.9 2.6
Hs7656849 28.5 3.9
YHL028w 27.7 5.5
> ECU10g0250
Length=1141
Score = 131 bits (329), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 62/114 (54%), Positives = 82/114 (71%), Gaps = 6/114 (5%)
Query 1 NLTYSSSLFVSIEQTTYKIDPLTNEEIVEERHSYERVCLGRLPIMLKSFYCWTHDISAEQ 60
+LTY+S LF+ + + T +E V ++H Y RV G LP+ML+S YC + + +
Sbjct 99 DLTYASPLFIDVTKETL------SELGVVDKHKYSRVPFGSLPVMLRSSYCVLYGLGDKD 152
Query 61 LPDVGECSYDQGGYFIINGSEKVLVAQERMASNFVYVFKKAQPSKFSWVAEVRS 114
L D+GEC YDQGGYFI+NGSEKV+VAQERMASN VYVFKKAQP+ ++ AE+RS
Sbjct 153 LIDLGECPYDQGGYFIVNGSEKVIVAQERMASNTVYVFKKAQPATYTHYAEIRS 206
> At4g21710
Length=1188
Score = 125 bits (313), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 64/120 (53%), Positives = 85/120 (70%), Gaps = 3/120 (2%)
Query 1 NLTYSSSLFVSIEQTTYKIDPLTNEEIVEERHSYERVCLGRLPIMLKSFYCWTHDISAEQ 60
NLTYS+ L+V + T I + E V E + +V +G++PIML+S YC S +
Sbjct 119 NLTYSAPLYVDV--TKRVIKKGHDGEEVTETQDFTKVFIGKVPIMLRSSYCTLFQNSEKD 176
Query 61 LPDVGECSYDQGGYFIINGSEKVLVAQERMASNFVYVFKKAQPSKFSWVAEVRSQMQGNQ 120
L ++GEC YDQGGYFIINGSEKVL+AQE+M++N VYVFKK QP+K+++V EVRS M NQ
Sbjct 177 LTELGECPYDQGGYFIINGSEKVLIAQEKMSTNHVYVFKKRQPNKYAYVGEVRS-MAENQ 235
> YOR151c
Length=1224
Score = 116 bits (291), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 68/137 (49%), Positives = 87/137 (63%), Gaps = 9/137 (6%)
Query 1 NLTYSSSLFVSIEQTTYK-ID----PLTNEEIVEERHSYE---RVCLGRLPIMLKSFYCW 52
NLTYSS LFV +++ TY+ ID L E I EE +V +GRLPIML+S C+
Sbjct 121 NLTYSSGLFVDVKKRTYEAIDVPGRELKYELIAEESEDDSESGKVFIGRLPIMLRSKNCY 180
Query 53 THDISAEQLPDVGECSYDQGGYFIINGSEKVLVAQERMASNFVYVFKKAQPSKFSWVAEV 112
+ + L + EC +D GGYFIINGSEKVL+AQER A N V VFKKA PS S VAE+
Sbjct 181 LSEATESDLYKLKECPFDMGGYFIINGSEKVLIAQERSAGNIVQVFKKAAPSPISHVAEI 240
Query 113 RSQMQ-GNQATSGFAIK 128
RS ++ G++ S +K
Sbjct 241 RSALEKGSRFISTLQVK 257
> SPAC23G3.01
Length=728
Score = 112 bits (281), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 63/136 (46%), Positives = 88/136 (64%), Gaps = 9/136 (6%)
Query 1 NLTYSSSLFVSIEQTTYKIDPLTNEEI-------VEERHSYERVCLGRLPIMLKSFYCWT 53
NLTYSS L+V + + + +N I EE +V +G++PIML+S +C
Sbjct 108 NLTYSSPLYVDMRKKVM-VAADSNVPIGEEEWLVEEEDEEPSKVFIGKIPIMLRSTFCIL 166
Query 54 HDISAEQLPDVGECSYDQGGYFIINGSEKVLVAQERMASNFVYVFKKAQPSKFSWVAEVR 113
+ +S +L D+ EC YDQGGYFIINGSEKV++AQER A+N V VFKKA PS ++VAE+R
Sbjct 167 NGVSDSELYDLNECPYDQGGYFIINGSEKVIIAQERSAANIVQVFKKAAPSPIAYVAEIR 226
Query 114 SQMQ-GNQATSGFAIK 128
S ++ G++ S IK
Sbjct 227 SALERGSRLISSMQIK 242
> Hs4505941
Length=1174
Score = 105 bits (262), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 54/119 (45%), Positives = 81/119 (68%), Gaps = 7/119 (5%)
Query 1 NLTYSSSLFVSIEQTTYKIDPLTNEEIVEERHSYERVCLGRLPIMLKSFYCWTHDISAEQ 60
NLTYS+ L+V I +T K EE ++ +H ++ +G++PIML+S YC + ++
Sbjct 117 NLTYSAPLYVDITKTVIK----EGEEQLQTQH--QKTFIGKIPIMLRSTYCLLNGLTDRD 170
Query 61 LPDVGECSYDQGGYFIINGSEKVLVAQERMASNFVYVFKKAQPSKFSWVAEVRSQMQGN 119
L ++ EC D GGYFIINGSEKVL+AQE+MA+N VYVF K + SK+++ E RS ++ +
Sbjct 171 LCELNECPLDPGGYFIINGSEKVLIAQEKMATNTVYVFAK-KDSKYAYTGECRSCLENS 228
> 7299847
Length=1176
Score = 100 bits (249), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 53/119 (44%), Positives = 78/119 (65%), Gaps = 11/119 (9%)
Query 1 NLTYSSSLFVSIEQT--TYKIDPLTNEEIVEERHSYERVCLGRLPIMLKSFYCWTHDISA 58
NLTYS+ L+V I +T +DP VE +H ++ +G++PIML+S YC ++
Sbjct 118 NLTYSAPLYVDITKTKNVEGLDP------VETQH--QKTFIGKIPIMLRSTYCLLSQLTD 169
Query 59 EQLPDVGECSYDQGGYFIINGSEKVLVAQERMASNFVYVFKKAQPSKFSWVAEVRSQMQ 117
L ++ EC D GGYFIINGSEKVL+AQE+MA+N VYVF + K+++ E+RS ++
Sbjct 170 RDLTELNECPLDPGGYFIINGSEKVLIAQEKMATNTVYVF-SMKDGKYAFKTEIRSCLE 227
> CE01162
Length=1193
Score = 100 bits (248), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 49/117 (41%), Positives = 76/117 (64%), Gaps = 8/117 (6%)
Query 1 NLTYSSSLFVSIEQTTYKIDPLTNEEIVEERHSYERVCLGRLPIMLKSFYCWTHDISAEQ 60
NLTY+S L+V I + + D T + Y++V +G++P+ML+S YC +++
Sbjct 124 NLTYASPLYVDITKVVTRDDSATEK-------VYDKVFVGKVPVMLRSSYCMLSNMTDRD 176
Query 61 LPDVGECSYDQGGYFIINGSEKVLVAQERMASNFVYVFKKAQPSKFSWVAEVRSQMQ 117
L ++ EC D GGYF+INGSEKVL+AQE+MA+N VYVF + K+++ E RS ++
Sbjct 177 LTELNECPLDPGGYFVINGSEKVLIAQEKMATNTVYVF-SMKDGKYAFKTECRSCLE 232
> At5g45140
Length=1194
Score = 75.9 bits (185), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 36/97 (37%), Positives = 59/97 (60%), Gaps = 7/97 (7%)
Query 1 NLTYSSSLFVSIEQTTYKIDPLTNEEIVEERHSYERVCLGRLPIMLKSFYCWTHDISAEQ 60
++TY++ +FV+IE + + + + + V +GR+PIML+S C H E+
Sbjct 132 DMTYAAPIFVNIEY-------VHGSHGNKAKSAKDNVIIGRMPIMLRSCRCVLHGKDEEE 184
Query 61 LPDVGECSYDQGGYFIINGSEKVLVAQERMASNFVYV 97
L +GEC D GGYFII G+EKVL+ QE+++ N + +
Sbjct 185 LARLGECPLDPGGYFIIKGTEKVLLIQEQLSKNRIII 221
> SPAC4G9.08c
Length=1165
Score = 75.1 bits (183), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 41/114 (35%), Positives = 67/114 (58%), Gaps = 13/114 (11%)
Query 1 NLTYSSSLFVSIEQTTYKIDPLTNEEIVEERHSYERVCLGRLPIMLKSFYCWTHDISAEQ 60
+LTY ++++V IE T K ++V R+ V +GR+P+ML+S C + +
Sbjct 130 DLTYGANIYVDIEYTRGK-------QVVRRRN----VPIGRMPVMLRSNKCVLSGKNEME 178
Query 61 LPDVGECSYDQGGYFIINGSEKVLVAQERMASNFVYVFKKAQPSKFSWVAEVRS 114
+ + EC D GGYFI+ G+EKV++ QE+++ N + V +A+P K W A V S
Sbjct 179 MAALNECPLDPGGYFIVKGTEKVILVQEQLSKNRIIV--EAEPKKGLWQASVTS 230
> CE00690
Length=1207
Score = 74.7 bits (182), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 43/137 (31%), Positives = 75/137 (54%), Gaps = 15/137 (10%)
Query 1 NLTYSSSLFVSIEQTTYKIDPLTNEEIVEERHSYERVCLGRLPIMLKSFYCWTHDISAEQ 60
++TYS+ + V IE T N+ + ++ + +GR+PIML+S C D++ E+
Sbjct 162 DMTYSAPISVDIEYTR------GNQRVFKKD-----LIIGRMPIMLRSSKCILRDLAEEE 210
Query 61 LPDVGECSYDQGGYFIINGSEKVLVAQERMASNFVYV----FKKAQPSKFSWVAEVRSQM 116
L V EC YD GGYF++ GSEKV++ QE+++ N + V K+ Q S +E +S+
Sbjct 211 LARVQECPYDPGGYFVVKGSEKVILIQEQLSKNRIMVGRNSSKELQCEVLSSTSERKSKT 270
Query 117 QGNQATSGFAIKKRTMT 133
++++ +T
Sbjct 271 YVTMKKGKYSVRHNQLT 287
> YOR207c
Length=1149
Score = 71.6 bits (174), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 35/97 (36%), Positives = 58/97 (59%), Gaps = 11/97 (11%)
Query 1 NLTYSSSLFVSIEQTTYKIDPLTNEEIVEERHSYERVCLGRLPIMLKSFYCWTHDISAEQ 60
++TYS+ ++V IE T I+ ++ V +GR+PIML+S C +D +
Sbjct 122 DMTYSAPIYVDIEYTR-------GRNII----MHKDVEIGRMPIMLRSNKCILYDADESK 170
Query 61 LPDVGECSYDQGGYFIINGSEKVLVAQERMASNFVYV 97
+ + EC D GGYFI+NG+EKV++ QE+++ N + V
Sbjct 171 MAKLNECPLDPGGYFIVNGTEKVILVQEQLSKNRIIV 207
> At3g18090
Length=1038
Score = 70.9 bits (172), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 39/120 (32%), Positives = 69/120 (57%), Gaps = 10/120 (8%)
Query 1 NLTYSSSLFVSIEQTTYKIDPLTNEEIVEERHSYERVCLGRLPIMLKSFYCWTHDISAEQ 60
N+TYS+ + V+ + T + D +EI++ + + + +G +P+M+KS C T + E
Sbjct 7 NMTYSARIKVNAQVETGQ-DEYVEKEILDVKK--QDILIGSIPVMVKSVLCKTSEKGKEN 63
Query 61 LPDVGECSYDQGGYFIINGSEKVLVAQERMASNFVYVFKKAQPSKFSWVAEVRSQMQGNQ 120
G C++DQGGYF+I G+EKV +AQE+M + +++ S W RS+ + N+
Sbjct 64 CKK-GNCAFDQGGYFVIKGAEKVFIAQEQMCTKRLWI------SNSPWTVSFRSETKRNR 116
> Hs22058718
Length=273
Score = 60.5 bits (145), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 38/129 (29%), Positives = 63/129 (48%), Gaps = 14/129 (10%)
Query 1 NLTYSSSLFVSIEQTTYKIDPLTNEEIVEERHSYERVCLGRLPIMLKSFYCWTHDISAEQ 60
++TYS+ + V IE T +R + +GR+PIML+S C + +
Sbjct 106 DMTYSAPITVDIEYTRG-----------SQRIIRNALPIGRMPIMLRSSNCVLTGKTPAE 154
Query 61 LPDVGECSYDQGGYFIINGSEKVLVAQERMASNFVYVFKKAQPSKFSWVAEVRSQMQGNQ 120
+ EC D GGYFI+ G EKV++ QE+++ N + V K + A V S +
Sbjct 155 FAKLNECPLDPGGYFIVKGVEKVILIQEQLSKNRIIV---EADRKGAVGASVTSSTHEKK 211
Query 121 ATSGFAIKK 129
+ + A+K+
Sbjct 212 SRTNMAVKQ 220
> ECU11g0370
Length=1110
Score = 58.2 bits (139), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 31/107 (28%), Positives = 58/107 (54%), Gaps = 21/107 (19%)
Query 1 NLTYSSSLFVSIEQTTYKIDPLTNEEIVEERHSYERVCLGRLPIMLKSFYCWTH------ 54
++TYS++++V IE + N +I+ +R VC+G++P+ML+S C
Sbjct 85 DITYSANIYVDIEY-------VRNRQIIVKRD----VCIGKMPVMLRSSRCHLRRETKGP 133
Query 55 ----DISAEQLPDVGECSYDQGGYFIINGSEKVLVAQERMASNFVYV 97
++ + EC D GGYFI+ G E+V++ QE+++ N + +
Sbjct 134 MEGRKTKDRRIRESQECPLDVGGYFIVRGIERVVLIQEQLSKNRIII 180
> 7303535
Length=1137
Score = 55.1 bits (131), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 48/93 (51%), Gaps = 11/93 (11%)
Query 1 NLTYSSSLFVSIEQTTYKIDPLTNEEIVEERHSYERVCLGRLPIMLKSFYCWTHDISAEQ 60
+ TYS+ + V IE T +R + +GR+P+ML+ C S +
Sbjct 114 DTTYSAPITVDIEYTRGT-----------QRIKRNNLLIGRMPLMLRCSNCALTGKSEFE 162
Query 61 LPDVGECSYDQGGYFIINGSEKVLVAQERMASN 93
L + EC D GGYF++ G EKV++ QE+++ N
Sbjct 163 LSKLNECPLDPGGYFVVRGQEKVILIQEQLSWN 195
> CE05629
Length=1127
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 34/92 (36%), Positives = 51/92 (55%), Gaps = 14/92 (15%)
Query 1 NLTYSSSLFVSIEQTTYKIDPLTNEEIVEERHSYERVCLGRLPIMLKSFYCWTHDISAEQ 60
LTY+ +L V I+ + +I+E + LG++PIML+S C +S ++
Sbjct 77 GLTYAGNLKVGIDVHVNG----SRLDIIE-------IILGKVPIMLRSEGCHLRGMSRKE 125
Query 61 LPDVGECSYDQGGYFIINGSEKV---LVAQER 89
L GE ++GGYFI+NGSEKV L+A R
Sbjct 126 LVVAGEEPIEKGGYFIVNGSEKVIRLLIANRR 157
> Hs9506619
Length=336
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/46 (45%), Positives = 30/46 (65%), Gaps = 0/46 (0%)
Query 39 LGRLPIMLKSFYCWTHDISAEQLPDVGECSYDQGGYFIINGSEKVL 84
LG +PIM+KS C ++ + L + E + + GGYFIING EKV+
Sbjct 134 LGYVPIMVKSKLCNLRNLPPQALIEHHEEAEEMGGYFIINGIEKVI 179
> ECU03g0440
Length=1062
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 41/83 (49%), Gaps = 7/83 (8%)
Query 24 NEEIVEERHSYERVCLGRLPIMLKSFYCWTHDISAEQLPDVGECSYDQGGYFIINGSEKV 83
N+ +V E S G PIM++S C + I + +VGE + GGYFIING ++
Sbjct 102 NKHVVTEDKS-----AGHFPIMVRSTLCHLNGIKNKN--EVGEDEDEIGGYFIINGIDRF 154
Query 84 LVAQERMASNFVYVFKKAQPSKF 106
+ N ++ FK + + F
Sbjct 155 IRFNIMQKRNHIFAFKMRKKTSF 177
> YPR010c
Length=1203
Score = 41.2 bits (95), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 25/63 (39%), Positives = 36/63 (57%), Gaps = 3/63 (4%)
Query 30 ERHSYERVCLGRLPIMLKSFYCWTHDISAEQLPDVGECSYDQGGYFIINGSEKV---LVA 86
E + +E G LP+ML+S C + +S +L E S + GGYFI+NG EK+ L+
Sbjct 149 EENLFEVRDCGGLPVMLQSNRCHLNKMSPYELVQHKEESDEIGGYFIVNGIEKLIRMLIV 208
Query 87 QER 89
Q R
Sbjct 209 QRR 211
> SPBP23A10.07
Length=1227
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 29/46 (63%), Gaps = 0/46 (0%)
Query 39 LGRLPIMLKSFYCWTHDISAEQLPDVGECSYDQGGYFIINGSEKVL 84
+G +PIM++S C +S +L E S + GGYFI+NG EK++
Sbjct 190 VGMIPIMVRSNRCHLEGLSPAELIAHKEESEEMGGYFIVNGIEKLI 235
> 7296211
Length=1129
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 28/46 (60%), Gaps = 0/46 (0%)
Query 39 LGRLPIMLKSFYCWTHDISAEQLPDVGECSYDQGGYFIINGSEKVL 84
LG +PIML+S C + E++ GE + GG F+I G+EK++
Sbjct 133 LGEVPIMLRSKACNLGQATPEEMVKHGEHDSEWGGIFVIRGNEKIV 178
> At1g29940
Length=1114
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 24/46 (52%), Gaps = 0/46 (0%)
Query 39 LGRLPIMLKSFYCWTHDISAEQLPDVGECSYDQGGYFIINGSEKVL 84
G+ PIML S C +L E + + GGYFI+NG E+V
Sbjct 71 FGQFPIMLMSKLCSLKGADCRKLLKCKESTSEMGGYFILNGIERVF 116
> 7299737
Length=1169
Score = 32.0 bits (71), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 23/43 (53%), Gaps = 6/43 (13%)
Query 19 IDPLTNEEIVEERHSYERVCLGRLPIMLKSFYCWTHDISAEQL 61
+DP N+E H + RVC RLPI K FY HD S + +
Sbjct 736 LDPDANDE----NHCHCRVCDSRLPI--KVFYLRQHDASRKHV 772
> CE01204
Length=425
Score = 32.0 bits (71), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 28/62 (45%), Gaps = 4/62 (6%)
Query 67 CSYDQGGYFIINGSEKVLVAQERMASNFVYVFKKAQPSKFSWVAE----VRSQMQGNQAT 122
C Y QG + S+KVL + R S V K + FS V E VR +Q N T
Sbjct 20 CEYSQGEKSVFMPSKKVLFEKYRRMSEIAEVVPKIIEAIFSSVQEPFKTVRPSLQHNHKT 79
Query 123 SG 124
SG
Sbjct 80 SG 81
> At5g60170_1
Length=533
Score = 30.4 bits (67), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 33/60 (55%), Gaps = 13/60 (21%)
Query 60 QLPDVGECSYDQGGYFIINGSEKVLVAQERMASNFVYVFKKAQPSKFSWVAEVRSQMQGN 119
+LPD+ +C D IN +K + ++R+ + +P + W+++++SQMQG+
Sbjct 455 KLPDLEQCRIDSS----INTDKKAISLEDRIP--------RTRPG-WDWISDLQSQMQGS 501
> Hs4758762
Length=548
Score = 28.9 bits (63), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 24/102 (23%), Positives = 42/102 (41%), Gaps = 19/102 (18%)
Query 35 ERVCLGRLPIMLKSFY---CWTHDISAEQ----LPDVGECSYDQGGYFIINGSEKVLVAQ 87
E + L R P+ +KSFY C E +P+VGE I+ GS ++ ++
Sbjct 433 EPILLCRFPVEIKSFYMQRCPEDSRLTESVDVLMPNVGE---------IVGGSMRIFDSE 483
Query 88 ERMASNFVYVFKKAQPSKFSWVAEVRSQMQGNQATSGFAIKK 129
E +A Y + P+ + W + R G +++
Sbjct 484 EILAG---YKREGIDPTPYYWYTDQRKYGTCPHGGYGLGLER 522
> Hs7656849
Length=415
Score = 28.5 bits (62), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 24/101 (23%), Positives = 44/101 (43%), Gaps = 8/101 (7%)
Query 5 SSSLFVSIEQ-TTYKIDPLTNEEIVEERHSYERVCLGRLPIMLKSFYCWTHDISAEQLPD 63
S++ ++SI + ID NEE +RH + C + + YC T S
Sbjct 118 STTAYISIHNMCAWMIDSFGNEE---QRHKF---CPPLCTMEKFASYCLTEPGSGSDAAS 171
Query 64 VGECSYDQGGYFIINGSEKVLVAQERMASNFVYVFKKAQPS 104
+ + QG ++I+NGS K ++ + +V + + P
Sbjct 172 LLTSAKKQGDHYILNGS-KAFISGAGESDIYVVMCRTGGPG 211
> YHL028w
Length=605
Score = 27.7 bits (60), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 12/37 (32%), Positives = 16/37 (43%), Gaps = 0/37 (0%)
Query 38 CLGRLPIMLKSFYCWTHDISAEQLPDVGECSYDQGGY 74
C G +++ F CW D VG+CS GY
Sbjct 56 CAGHQFAIVQGFMCWCSDSEPSTQTSVGDCSGTCPGY 92
Lambda K H
0.318 0.131 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1356426142
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40