bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1287_orf2
Length=120
Score E
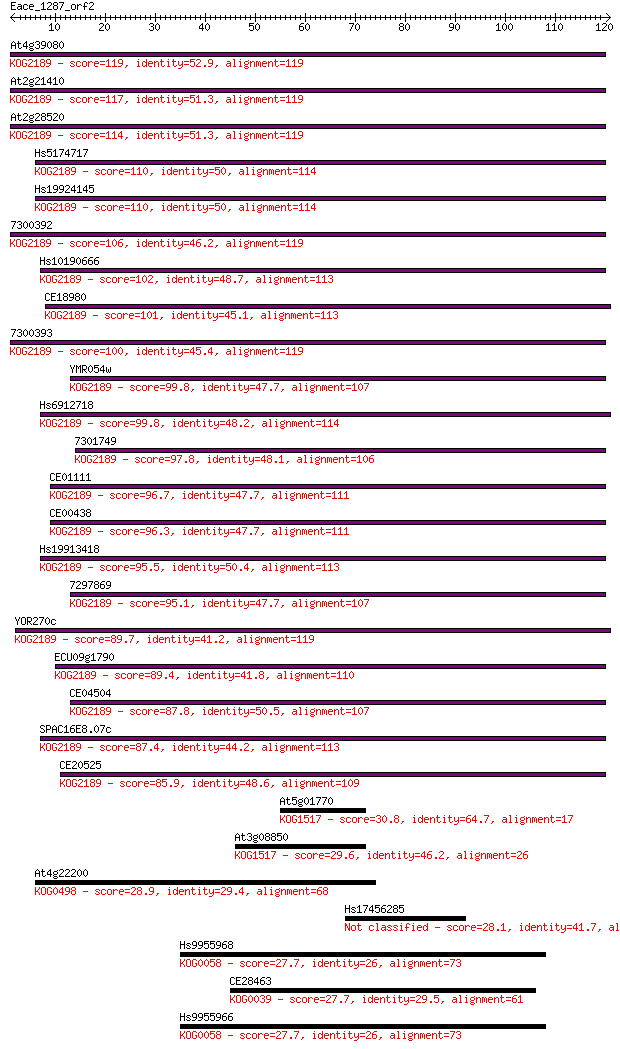
Sequences producing significant alignments: (Bits) Value
At4g39080 119 1e-27
At2g21410 117 7e-27
At2g28520 114 4e-26
Hs5174717 110 5e-25
Hs19924145 110 5e-25
7300392 106 1e-23
Hs10190666 102 2e-22
CE18980 101 4e-22
7300393 100 5e-22
YMR054w 99.8 1e-21
Hs6912718 99.8 1e-21
7301749 97.8 4e-21
CE01111 96.7 9e-21
CE00438 96.3 1e-20
Hs19913418 95.5 2e-20
7297869 95.1 3e-20
YOR270c 89.7 1e-18
ECU09g1790 89.4 1e-18
CE04504 87.8 4e-18
SPAC16E8.07c 87.4 6e-18
CE20525 85.9 1e-17
At5g01770 30.8 0.62
At3g08850 29.6 1.6
At4g22200 28.9 2.2
Hs17456285 28.1 3.6
Hs9955968 27.7 5.9
CE28463 27.7 5.9
Hs9955966 27.7 5.9
> At4g39080
Length=843
Score = 119 bits (298), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 63/119 (52%), Positives = 84/119 (70%), Gaps = 5/119 (4%)
Query 1 GVAAAEEEMDVTELWLHNMIEGIEFVLGSISNTASYLRLWALSLAHQQLSCIFFEKTVGL 60
G + EE + +E+++H +I IEFVLG++SNTASYLRLWALSLAH +LS +F+EK + L
Sbjct 714 GGSHGHEEFEFSEIFVHQLIHTIEFVLGAVSNTASYLRLWALSLAHSELSSVFYEKVLLL 773
Query 61 AFQPGLSPSQMAFKLIFFFPAFLCITIFVMVAMEALECFLHALRLQWVEFQNKFYKGDG 119
A+ G + + LI F+ T+ V++ ME L FLHALRL WVEFQNKFY+GDG
Sbjct 774 AW--GYNNPLI---LIVGVLVFIFATVGVLLVMETLSAFLHALRLHWVEFQNKFYEGDG 827
> At2g21410
Length=821
Score = 117 bits (292), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 61/119 (51%), Positives = 82/119 (68%), Gaps = 5/119 (4%)
Query 1 GVAAAEEEMDVTELWLHNMIEGIEFVLGSISNTASYLRLWALSLAHQQLSCIFFEKTVGL 60
G EE + +E+++H +I IEFVLG++SNTASYLRLWALSLAH +LS +F+EK + +
Sbjct 692 GGGHGHEEFEFSEIFVHQLIHTIEFVLGAVSNTASYLRLWALSLAHSELSSVFYEKVLLM 751
Query 61 AFQPGLSPSQMAFKLIFFFPAFLCITIFVMVAMEALECFLHALRLQWVEFQNKFYKGDG 119
A+ G + F I F+ T+ V++ ME L FLHALRL WVE+QNKFY+GDG
Sbjct 752 AW--GFNN---VFIWIVGILVFIFATVGVLLVMETLSAFLHALRLHWVEYQNKFYEGDG 805
> At2g28520
Length=780
Score = 114 bits (285), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 61/119 (51%), Positives = 79/119 (66%), Gaps = 5/119 (4%)
Query 1 GVAAAEEEMDVTELWLHNMIEGIEFVLGSISNTASYLRLWALSLAHQQLSCIFFEKTVGL 60
G EEE + +E+++H +I IEFVLGS+SNTASYLRLWALSLAH +LS +F+EK + L
Sbjct 656 GGGHHEEEFNFSEIFVHQLIHSIEFVLGSVSNTASYLRLWALSLAHSELSTVFYEKVLLL 715
Query 61 AFQPGLSPSQMAFKLIFFFPAFLCITIFVMVAMEALECFLHALRLQWVEFQNKFYKGDG 119
A+ + +LI F T F+++ ME L FLHALRL WVEF KF+ GDG
Sbjct 716 AW----GYENILIRLI-GVAVFAFATAFILLMMETLSAFLHALRLHWVEFMGKFFNGDG 769
> Hs5174717
Length=614
Score = 110 bits (276), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 57/114 (50%), Positives = 72/114 (63%), Gaps = 0/114 (0%)
Query 6 EEEMDVTELWLHNMIEGIEFVLGSISNTASYLRLWALSLAHQQLSCIFFEKTVGLAFQPG 65
E E+ +E+ +H I IEF LG +SNTASYLRLWALSLAH QLS + + + + G
Sbjct 488 EAELVPSEVLMHQAIHTIEFCLGCVSNTASYLRLWALSLAHAQLSEVLWAMVMRIGLGLG 547
Query 66 LSPSQMAFKLIFFFPAFLCITIFVMVAMEALECFLHALRLQWVEFQNKFYKGDG 119
A L+ F AF +T+ +++ ME L FLHALRL WVEFQNKFY G G
Sbjct 548 REVGVAAVVLVPIFAAFAVMTVAILLVMEGLSAFLHALRLHWVEFQNKFYSGTG 601
> Hs19924145
Length=830
Score = 110 bits (276), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 57/114 (50%), Positives = 72/114 (63%), Gaps = 0/114 (0%)
Query 6 EEEMDVTELWLHNMIEGIEFVLGSISNTASYLRLWALSLAHQQLSCIFFEKTVGLAFQPG 65
E E+ +E+ +H I IEF LG +SNTASYLRLWALSLAH QLS + + + + G
Sbjct 704 EAELVPSEVLMHQAIHTIEFCLGCVSNTASYLRLWALSLAHAQLSEVLWAMVMRIGLGLG 763
Query 66 LSPSQMAFKLIFFFPAFLCITIFVMVAMEALECFLHALRLQWVEFQNKFYKGDG 119
A L+ F AF +T+ +++ ME L FLHALRL WVEFQNKFY G G
Sbjct 764 REVGVAAVVLVPIFAAFAVMTVAILLVMEGLSAFLHALRLHWVEFQNKFYSGTG 817
> 7300392
Length=844
Score = 106 bits (264), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 55/119 (46%), Positives = 76/119 (63%), Gaps = 2/119 (1%)
Query 1 GVAAAEEEMDVTELWLHNMIEGIEFVLGSISNTASYLRLWALSLAHQQLSCIFFEKTVGL 60
G + E++ ++E+W+H I IE++L +IS+TASYLRLWALSLAH QLS + + + +
Sbjct 710 GGHSEEDDEPMSEIWIHQAIHTIEYILSTISHTASYLRLWALSLAHAQLSEVLWTMVLAM 769
Query 61 AFQPGLSPSQMAFKLIFFFPAFLCITIFVMVAMEALECFLHALRLQWVEFQNKFYKGDG 119
Q ++ A L F F + TI +MV ME L FLH LRL WVEF +KFY G+G
Sbjct 770 GLQ--MNGYVGAIGLFFIFAVWEFFTIAIMVMMEGLSAFLHTLRLHWVEFMSKFYVGNG 826
> Hs10190666
Length=840
Score = 102 bits (253), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 55/113 (48%), Positives = 71/113 (62%), Gaps = 2/113 (1%)
Query 7 EEMDVTELWLHNMIEGIEFVLGSISNTASYLRLWALSLAHQQLSCIFFEKTVGLAFQPGL 66
EE + ++++H I IE+ LG ISNTASYLRLWALSLAH QLS + + + Q
Sbjct 712 EEFNFGDVFVHQAIHTIEYCLGCISNTASYLRLWALSLAHAQLSEVLWTMVMNSGLQTRG 771
Query 67 SPSQMAFKLIFFFPAFLCITIFVMVAMEALECFLHALRLQWVEFQNKFYKGDG 119
+ +IF F +T+ +++ ME L FLHALRL WVEFQNKFY GDG
Sbjct 772 WGGIVGVFIIF--AVFAVLTVAILLIMEGLSAFLHALRLHWVEFQNKFYVGDG 822
> CE18980
Length=865
Score = 101 bits (251), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 51/113 (45%), Positives = 74/113 (65%), Gaps = 1/113 (0%)
Query 8 EMDVTELWLHNMIEGIEFVLGSISNTASYLRLWALSLAHQQLSCIFFEKTVGLAFQPGLS 67
E + ++ +H I IEFVLG +S+TASYLRLWALSLAH QLS + + + ++ G
Sbjct 735 EFNFGDIMVHQAIHTIEFVLGCVSHTASYLRLWALSLAHAQLSDVLWTMVLRMSLTMGGW 794
Query 68 PSQMAFKLIFFFPAFLCITIFVMVAMEALECFLHALRLQWVEFQNKFYKGDGV 120
A ++F+F F +++ +++ ME L FLHA+RL WVEFQ+KFY G G+
Sbjct 795 GGSAAITILFYF-IFSILSVCILILMEGLSAFLHAIRLHWVEFQSKFYGGTGI 846
> 7300393
Length=815
Score = 100 bits (250), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 54/119 (45%), Positives = 76/119 (63%), Gaps = 2/119 (1%)
Query 1 GVAAAEEEMDVTELWLHNMIEGIEFVLGSISNTASYLRLWALSLAHQQLSCIFFEKTVGL 60
G A ++ ++E+++H I IE+VL +IS+TASYLRLWALSLAH QLS + ++ + L
Sbjct 681 GGAHGHDDEPMSEIYIHQAIHTIEYVLSTISHTASYLRLWALSLAHAQLSEVLWQMVLSL 740
Query 61 AFQPGLSPSQMAFKLIFFFPAFLCITIFVMVAMEALECFLHALRLQWVEFQNKFYKGDG 119
+ +S A L F A+ T+ ++V ME L FLH LRL WVEF +KFY+G G
Sbjct 741 GLK--MSGVGGAIGLFIIFGAWCLFTLAILVLMEGLSAFLHTLRLHWVEFMSKFYEGMG 797
> YMR054w
Length=890
Score = 99.8 bits (247), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 51/108 (47%), Positives = 72/108 (66%), Gaps = 1/108 (0%)
Query 13 ELWLHNMIEGIEFVLGSISNTASYLRLWALSLAHQQLSCIFFEKTVGLAFQPGLSPSQMA 72
++ +H +I IEF L IS+TASYLRLWALSLAH QLS + ++ T+ AF S S +A
Sbjct 770 DVMIHQVIHTIEFCLNCISHTASYLRLWALSLAHAQLSSVLWDMTISNAFSSKNSGSPLA 829
Query 73 -FKLIFFFPAFLCITIFVMVAMEALECFLHALRLQWVEFQNKFYKGDG 119
K++F F + +T+ ++V ME LHALRL WVE +KF++G+G
Sbjct 830 VMKVVFLFAMWFVLTVCILVFMEGTSAMLHALRLHWVEAMSKFFEGEG 877
> Hs6912718
Length=856
Score = 99.8 bits (247), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 55/117 (47%), Positives = 72/117 (61%), Gaps = 8/117 (6%)
Query 7 EEMDVTELWLHNMIEGIEFVLGSISNTASYLRLWALSLAHQQLSCIFFEKTVGLAFQPGL 66
EE + E+ + +I IE+ LG ISNTASYLRLWALSLAH QLS + + + + GL
Sbjct 724 EEFNFGEILMTQVIHSIEYCLGCISNTASYLRLWALSLAHAQLSDVLW----AMLMRVGL 779
Query 67 SPSQMAFKLIFFFPA---FLCITIFVMVAMEALECFLHALRLQWVEFQNKFYKGDGV 120
+ ++ P F +TIF+++ ME L FLHA+RL WVEFQNKFY G G
Sbjct 780 R-VDTTYGVLLLLPVIALFAVLTIFILLIMEGLSAFLHAIRLHWVEFQNKFYVGAGT 835
> 7301749
Length=855
Score = 97.8 bits (242), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 51/106 (48%), Positives = 68/106 (64%), Gaps = 2/106 (1%)
Query 14 LWLHNMIEGIEFVLGSISNTASYLRLWALSLAHQQLSCIFFEKTVGLAFQPGLSPSQMAF 73
+++H I IE+VLGS+S+TASYLRLWALSLAH QL+ + + + + + +
Sbjct 731 IFIHQSIHTIEYVLGSVSHTASYLRLWALSLAHAQLAEVLWTMVLSIGLKQEGPVGGIVL 790
Query 74 KLIFFFPAFLCITIFVMVAMEALECFLHALRLQWVEFQNKFYKGDG 119
+F F A L + I V+ ME L FLH LRL WVEFQ+KFYKG G
Sbjct 791 TCVFAFWAILTVGILVL--MEGLSAFLHTLRLHWVEFQSKFYKGQG 834
> CE01111
Length=1030
Score = 96.7 bits (239), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 53/111 (47%), Positives = 73/111 (65%), Gaps = 2/111 (1%)
Query 9 MDVTELWLHNMIEGIEFVLGSISNTASYLRLWALSLAHQQLSCIFFEKTVGLAFQPGLSP 68
+++ ++ ++ I IEFVLG +S+TASYLRLWALSLAH QLS + + AF
Sbjct 900 LEMGDVMVYQAIHTIEFVLGCVSHTASYLRLWALSLAHAQLSDVLWTMVFRNAFVLDGYT 959
Query 69 SQMAFKLIFFFPAFLCITIFVMVAMEALECFLHALRLQWVEFQNKFYKGDG 119
+A ++FF F +++F++V ME L FLHALRL WVEFQ+KFY G G
Sbjct 960 GAIATYILFFI--FGSLSVFILVLMEGLSAFLHALRLHWVEFQSKFYGGLG 1008
> CE00438
Length=935
Score = 96.3 bits (238), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 53/111 (47%), Positives = 73/111 (65%), Gaps = 2/111 (1%)
Query 9 MDVTELWLHNMIEGIEFVLGSISNTASYLRLWALSLAHQQLSCIFFEKTVGLAFQPGLSP 68
+++ ++ ++ I IEFVLG +S+TASYLRLWALSLAH QLS + + AF
Sbjct 805 LEMGDVMVYQAIHTIEFVLGCVSHTASYLRLWALSLAHAQLSDVLWTMVFRNAFVLDGYT 864
Query 69 SQMAFKLIFFFPAFLCITIFVMVAMEALECFLHALRLQWVEFQNKFYKGDG 119
+A ++FF F +++F++V ME L FLHALRL WVEFQ+KFY G G
Sbjct 865 GAIATYILFFI--FGSLSVFILVLMEGLSAFLHALRLHWVEFQSKFYGGLG 913
> Hs19913418
Length=831
Score = 95.5 bits (236), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 57/115 (49%), Positives = 71/115 (61%), Gaps = 6/115 (5%)
Query 7 EEMDVTELWLHNMIEGIEFVLGSISNTASYLRLWALSLAHQQLSCIFFEKTVGLAFQPGL 66
+E D + +H I IE+ LG ISNTASYLRLWALSLAH QLS + + + + GL
Sbjct 703 DEFDFGDTMVHQAIHTIEYCLGCISNTASYLRLWALSLAHAQLSEVLWTMVIHI----GL 758
Query 67 SPSQMAFKLIFFFPAFLCIT--IFVMVAMEALECFLHALRLQWVEFQNKFYKGDG 119
S +A L+ FF T + +++ ME L FLHALRL WVEFQNKFY G G
Sbjct 759 SVKSLAGGLVLFFFFTAFATLTVAILLIMEGLSAFLHALRLHWVEFQNKFYSGTG 813
> 7297869
Length=814
Score = 95.1 bits (235), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 51/108 (47%), Positives = 72/108 (66%), Gaps = 4/108 (3%)
Query 13 ELWLHNMIEGIEFVLGSISNTASYLRLWALSLAHQQLSCIFFEKTVGLAFQPGLSPSQMA 72
E+++H I IE+VL ++S+TASYLRLWALSLAH QLS + + + F+ S +
Sbjct 692 EIFIHQAIHTIEYVLSTVSHTASYLRLWALSLAHAQLSEVLWNMVFSMGFK---YDSYIG 748
Query 73 FKLIF-FFPAFLCITIFVMVAMEALECFLHALRLQWVEFQNKFYKGDG 119
LI+ FF A+ +T+ ++V +E L FLH LRL WVEF +KFY+G G
Sbjct 749 GILIYVFFGAWALLTVGILVLIEGLSAFLHTLRLHWVEFMSKFYEGAG 796
> YOR270c
Length=840
Score = 89.7 bits (221), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 49/119 (41%), Positives = 69/119 (57%), Gaps = 2/119 (1%)
Query 2 VAAAEEEMDVTELWLHNMIEGIEFVLGSISNTASYLRLWALSLAHQQLSCIFFEKTVGLA 61
V + D ++ +H +I IEF L +S+TASYLRLWALSLAH QLS + + T+ +A
Sbjct 699 VGSGSHGEDFGDIMIHQVIHTIEFCLNCVSHTASYLRLWALSLAHAQLSSVLWTMTIQIA 758
Query 62 FQPGLSPSQMAFKLIFFFPAFLCITIFVMVAMEALECFLHALRLQWVEFQNKFYKGDGV 120
F G F + F + +T V+V ME LH+LRL WVE +KF+ G+G+
Sbjct 759 F--GFRGFVGVFMTVALFAMWFALTCAVLVLMEGTSAMLHSLRLHWVESMSKFFVGEGL 815
> ECU09g1790
Length=700
Score = 89.4 bits (220), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 46/111 (41%), Positives = 72/111 (64%), Gaps = 11/111 (9%)
Query 10 DVTELWLHNMIEGIEFVLGSISNTASYLRLWALSLAHQQLSCIFFEKTVGLAFQPG-LSP 68
+++ LW++ I +EF LG ISNT+SYLRLWA+SLAH QL+ + E T+G + G ++P
Sbjct 585 EISSLWINQFIHVVEFGLGLISNTSSYLRLWAVSLAHAQLTRVLHEFTIG---KEGFIAP 641
Query 69 SQMAFKLIFFFPAFLCITIFVMVAMEALECFLHALRLQWVEFQNKFYKGDG 119
++ ++ T+ +++ ME L LHA+RL WVEF +KF++G G
Sbjct 642 VALS-------GVYVLGTVVLLIGMEGLGSCLHAMRLNWVEFHSKFFRGRG 685
> CE04504
Length=873
Score = 87.8 bits (216), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 54/110 (49%), Positives = 69/110 (62%), Gaps = 7/110 (6%)
Query 13 ELWLHNMIEGIEFVLGSISNTASYLRLWALSLAHQQLSCIFFEK---TVGLAFQPGLSPS 69
++ +H I IE+VLG +S+TASYLRLWALSLAH QLS + + T GL G+S +
Sbjct 750 DIMVHQAIHTIEYVLGCVSHTASYLRLWALSLAHAQLSEVLWHMVFVTGGL----GISGT 805
Query 70 QMAFKLIFFFPAFLCITIFVMVAMEALECFLHALRLQWVEFQNKFYKGDG 119
+ F F +TI ++V ME L FLH LRL WVEFQ+KFY G G
Sbjct 806 AGFIAVYVVFFIFFVLTISILVLMEGLSAFLHTLRLHWVEFQSKFYLGLG 855
> SPAC16E8.07c
Length=805
Score = 87.4 bits (215), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 50/114 (43%), Positives = 70/114 (61%), Gaps = 4/114 (3%)
Query 7 EEMDVTELWLHNMIEGIEFVLGSISNTASYLRLWALSLAHQQLSCIFFEKTVGLAFQ-PG 65
E ++ E+ +H +I IEF LG +S+TASYLRLWALSLAH QLS + + T+ F+ G
Sbjct 682 EPFELGEVVIHQVIHTIEFCLGCVSHTASYLRLWALSLAHNQLSSVLWNMTLANGFRMTG 741
Query 66 LSPSQMAFKLIFFFPAFLCITIFVMVAMEALECFLHALRLQWVEFQNKFYKGDG 119
+ S L F+ C+ V+VAME LH+LRL WVE +K ++G+G
Sbjct 742 IVGSIFVVILFGFWFIATCV---VLVAMEGTSAMLHSLRLHWVEGMSKHFEGEG 792
> CE20525
Length=1236
Score = 85.9 bits (211), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 53/142 (37%), Positives = 77/142 (54%), Gaps = 33/142 (23%)
Query 11 VTELWLHNMIEGIEFVLGSISNTASYLRLWALSLAHQQLSCIFF---------------E 55
++++++H I IEFVLG +S+TASYLRLWALSLAH QLS + +
Sbjct 1059 LSDIFVHQAIHTIEFVLGCVSHTASYLRLWALSLAHAQLSEVMWHMVLIQGIHTVDHIEN 1118
Query 56 KTVGLAFQPGLS-PSQMAFKLIFFFP-----------------AFLCITIFVMVAMEALE 97
+T+ + +P ++ + IFFF F +++ +++ ME L
Sbjct 1119 ETIAMCLKPVVACVGYFSASAIFFFCLTSLLYGKTYEKEKAFFIFASLSLSILIMMEGLS 1178
Query 98 CFLHALRLQWVEFQNKFYKGDG 119
FLHALRL WVEFQ+KFY G G
Sbjct 1179 AFLHALRLHWVEFQSKFYLGTG 1200
> At5g01770
Length=1354
Score = 30.8 bits (68), Expect = 0.62, Method: Composition-based stats.
Identities = 11/17 (64%), Positives = 15/17 (88%), Gaps = 0/17 (0%)
Query 55 EKTVGLAFQPGLSPSQM 71
EK VGL+FQPGL P+++
Sbjct 1226 EKVVGLSFQPGLDPAKI 1242
> At3g08850
Length=1384
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 12/26 (46%), Positives = 19/26 (73%), Gaps = 5/26 (19%)
Query 46 HQQLSCIFFEKTVGLAFQPGLSPSQM 71
HQ++ E+ VGL+FQPGL P+++
Sbjct 1253 HQKV-----ERVVGLSFQPGLDPAKV 1273
> At4g22200
Length=787
Score = 28.9 bits (63), Expect = 2.2, Method: Composition-based stats.
Identities = 20/70 (28%), Positives = 35/70 (50%), Gaps = 2/70 (2%)
Query 6 EEEMDVTELWLHNMIEGIEFVLGSISNTASYLRL--WALSLAHQQLSCIFFEKTVGLAFQ 63
EEE D + L L+N+ + I LG S +++R W +S + C F + +A+
Sbjct 15 EEEYDASSLSLNNLSKLILPPLGVASYNQNHIRSSGWIISPMDSRYRCWEFYMVLLVAYS 74
Query 64 PGLSPSQMAF 73
+ P ++AF
Sbjct 75 AWVYPFEVAF 84
> Hs17456285
Length=221
Score = 28.1 bits (61), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 10/24 (41%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 68 PSQMAFKLIFFFPAFLCITIFVMV 91
P+Q +L F+ P +LC+TIF ++
Sbjct 56 PTQTPEELAFYAPNYLCLTIFAIL 79
> Hs9955968
Length=723
Score = 27.7 bits (60), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 29/73 (39%), Gaps = 3/73 (4%)
Query 35 SYLRLWALSLAHQQLSCIFFEKTVGLAFQPGLSPSQMAFKLIFFFPAFLCITIFVMVAME 94
S L LWA L SC+ T+G+A L P ++ + L + I+ MV +
Sbjct 46 SVLDLWAACLYR---SCLLLGATIGVAKNSALGPRRLRASWLVITLVCLFVGIYAMVKLL 102
Query 95 ALECFLHALRLQW 107
+R W
Sbjct 103 LFSEVRRPIRDPW 115
> CE28463
Length=1497
Score = 27.7 bits (60), Expect = 5.9, Method: Composition-based stats.
Identities = 18/65 (27%), Positives = 29/65 (44%), Gaps = 6/65 (9%)
Query 45 AHQQLSCIFFEKTVGLAFQPGLSPSQMAFKLIFFFPAFLCITIFVMVAMEALECFL---- 100
+ + L+C+F E G F P + S F I + + ++ + AL CF+
Sbjct 1102 SQEGLACLFQEAFFGSNFLPSI--SYWFFSTITGLTGIALVAVMCIIYVFALPCFIKRAY 1159
Query 101 HALRL 105
HA RL
Sbjct 1160 HAFRL 1164
> Hs9955966
Length=766
Score = 27.7 bits (60), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 29/73 (39%), Gaps = 3/73 (4%)
Query 35 SYLRLWALSLAHQQLSCIFFEKTVGLAFQPGLSPSQMAFKLIFFFPAFLCITIFVMVAME 94
S L LWA L SC+ T+G+A L P ++ + L + I+ MV +
Sbjct 46 SVLDLWAACLYR---SCLLLGATIGVAKNSALGPRRLRASWLVITLVCLFVGIYAMVKLL 102
Query 95 ALECFLHALRLQW 107
+R W
Sbjct 103 LFSEVRRPIRDPW 115
Lambda K H
0.330 0.140 0.440
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160968786
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40