bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1265_orf2
Length=108
Score E
Sequences producing significant alignments: (Bits) Value
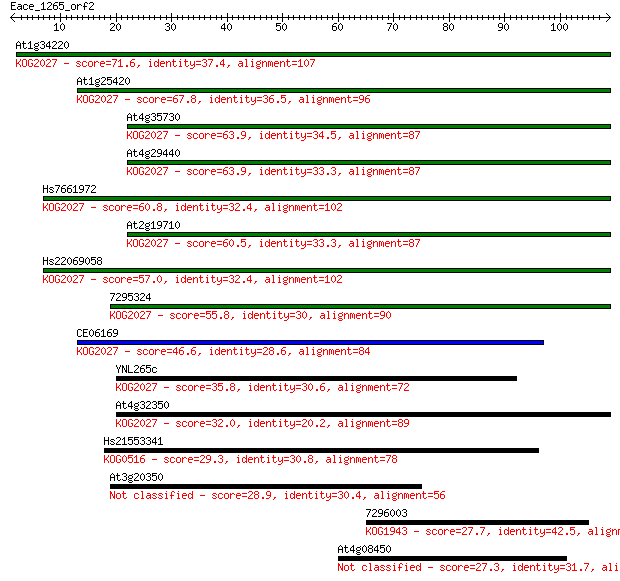
At1g34220 71.6 3e-13
At1g25420 67.8 5e-12
At4g35730 63.9 6e-11
At4g29440 63.9 7e-11
Hs7661972 60.8 5e-10
At2g19710 60.5 7e-10
Hs22069058 57.0 9e-09
7295324 55.8 2e-08
CE06169 46.6 1e-05
YNL265c 35.8 0.018
At4g32350 32.0 0.28
Hs21553341 29.3 2.0
At3g20350 28.9 2.1
7296003 27.7 5.4
At4g08450 27.3 6.3
> At1g34220
Length=619
Score = 71.6 bits (174), Expect = 3e-13, Method: Composition-based stats.
Identities = 40/107 (37%), Positives = 59/107 (55%), Gaps = 9/107 (8%)
Query 2 WKATKLATLFHGRRWPRSLKMAISRARMCTNRLQNSIRLQRKEIAGFLREGREEGARLKG 61
+KA K TL LK+ I R ++ NR + I+ R+EIA L G+E AR++
Sbjct 12 FKAAKCKTL---------LKLTIPRIKLIRNRREAQIKQMRREIAKLLETGQEATARIRV 62
Query 62 EHLLREYRLERAMEILVTVCELLISRLSYLATERTCPPDLDSPIHTL 108
EH++RE ++ A EIL CEL+ RL + +R CP DL I ++
Sbjct 63 EHIIREEKMMAAQEILELFCELIAVRLPIIEAQRECPLDLKEAISSV 109
> At1g25420
Length=323
Score = 67.8 bits (164), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 35/96 (36%), Positives = 59/96 (61%), Gaps = 0/96 (0%)
Query 13 GRRWPRSLKMAISRARMCTNRLQNSIRLQRKEIAGFLREGREEGARLKGEHLLREYRLER 72
G + SL +AI+R ++ N+ ++ +KEIA FL+ G+E AR++ EH++RE L
Sbjct 14 GAKCKTSLNLAIARMKLLQNKRDMQLKHMKKEIAHFLQAGQEPIARIRVEHVIREMNLWA 73
Query 73 AMEILVTVCELLISRLSYLATERTCPPDLDSPIHTL 108
A EIL CE +++R+ L +E+ CP +L I ++
Sbjct 74 AYEILELFCEFILARVPILESEKECPRELREAIASI 109
> At4g35730
Length=430
Score = 63.9 bits (154), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 30/87 (34%), Positives = 56/87 (64%), Gaps = 0/87 (0%)
Query 22 MAISRARMCTNRLQNSIRLQRKEIAGFLREGREEGARLKGEHLLREYRLERAMEILVTVC 81
MA++R ++ N+ ++ R++IA L+ G++ AR++ EH++RE ++ A EI+ C
Sbjct 1 MAVARIKLIRNKRLVVVKQMRRDIAVLLQSGQDATARIRVEHVIREQNIQAANEIIELFC 60
Query 82 ELLISRLSYLATERTCPPDLDSPIHTL 108
EL++SRL+ + ++ CP DL I +L
Sbjct 61 ELIVSRLTIITKQKQCPVDLKEGIASL 87
> At4g29440
Length=1071
Score = 63.9 bits (154), Expect = 7e-11, Method: Composition-based stats.
Identities = 29/87 (33%), Positives = 54/87 (62%), Gaps = 0/87 (0%)
Query 22 MAISRARMCTNRLQNSIRLQRKEIAGFLREGREEGARLKGEHLLREYRLERAMEILVTVC 81
MA SR ++ N+ I+ R+E+A L G+ + A+++ EH++RE + A E++ C
Sbjct 1 MAASRLKILKNKKDTQIKQLRRELAHLLESGQTQTAKIRVEHVVREEKTVAAYELVGIYC 60
Query 82 ELLISRLSYLATERTCPPDLDSPIHTL 108
ELL++RL + +++TCP DL + ++
Sbjct 61 ELLVARLGVIDSQKTCPNDLKEAVASV 87
> Hs7661972
Length=364
Score = 60.8 bits (146), Expect = 5e-10, Method: Composition-based stats.
Identities = 33/102 (32%), Positives = 56/102 (54%), Gaps = 0/102 (0%)
Query 7 LATLFHGRRWPRSLKMAISRARMCTNRLQNSIRLQRKEIAGFLREGREEGARLKGEHLLR 66
L + F R +L++ I+R ++ + + RKEIA +L G++E AR++ EH++R
Sbjct 2 LGSGFKAERLRVNLRLVINRLKLLEKKKTELAQKARKEIADYLAAGKDERARIRVEHIIR 61
Query 67 EYRLERAMEILVTVCELLISRLSYLATERTCPPDLDSPIHTL 108
E L AMEIL C+LL++R + + + L + TL
Sbjct 62 EDYLVEAMEILELYCDLLLARFGLIQSMKELDSGLAESVSTL 103
> At2g19710
Length=918
Score = 60.5 bits (145), Expect = 7e-10, Method: Composition-based stats.
Identities = 29/87 (33%), Positives = 52/87 (59%), Gaps = 0/87 (0%)
Query 22 MAISRARMCTNRLQNSIRLQRKEIAGFLREGREEGARLKGEHLLREYRLERAMEILVTVC 81
MA SR ++ N+ + I+ R+E+A L G+ AR++ EH++RE + A E++ C
Sbjct 1 MANSRLKILKNKKEIQIKQLRRELAQLLESGQTPTARIRVEHVVREEKTVAAYELIGIYC 60
Query 82 ELLISRLSYLATERTCPPDLDSPIHTL 108
ELL+ RL + +++ CP DL + ++
Sbjct 61 ELLVVRLGVIESQKNCPIDLKEAVTSV 87
> Hs22069058
Length=360
Score = 57.0 bits (136), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 33/102 (32%), Positives = 56/102 (54%), Gaps = 0/102 (0%)
Query 7 LATLFHGRRWPRSLKMAISRARMCTNRLQNSIRLQRKEIAGFLREGREEGARLKGEHLLR 66
L + F R +L++ I+R ++ + + RKEIA +L G++E AR++ EH++R
Sbjct 2 LGSGFKAERLRVNLRLVINRLKLLEKKKTELAQKARKEIADYLAAGKDERARIRVEHIIR 61
Query 67 EYRLERAMEILVTVCELLISRLSYLATERTCPPDLDSPIHTL 108
E L AMEIL C+LL++R + + + L + TL
Sbjct 62 EDYLVEAMEILELYCDLLLARFGLIQSMKELDSGLAESVSTL 103
> 7295324
Length=417
Score = 55.8 bits (133), Expect = 2e-08, Method: Composition-based stats.
Identities = 27/90 (30%), Positives = 52/90 (57%), Gaps = 0/90 (0%)
Query 19 SLKMAISRARMCTNRLQNSIRLQRKEIAGFLREGREEGARLKGEHLLREYRLERAMEILV 78
+L++A++R ++ + + RKEIA +L G+ E AR++ EH++RE L AME++
Sbjct 14 NLRLALNRLKLLEKKKAELTQKSRKEIADYLATGKTERARIRVEHIIREDYLVEAMEMVE 73
Query 79 TVCELLISRLSYLATERTCPPDLDSPIHTL 108
C+LL++R + + + P+ +L
Sbjct 74 MYCDLLLARFGLITQMKELDTGIAEPVASL 103
> CE06169
Length=432
Score = 46.6 bits (109), Expect = 1e-05, Method: Composition-based stats.
Identities = 24/87 (27%), Positives = 48/87 (55%), Gaps = 3/87 (3%)
Query 13 GRRWPR---SLKMAISRARMCTNRLQNSIRLQRKEIAGFLREGREEGARLKGEHLLREYR 69
G ++P+ +L++ I+R ++ + R EIA ++ + + AR++ EH++RE
Sbjct 6 GAQYPKLKTNLRLGINRLQLLGKKKTEMAMKARTEIADYIAANKPDRARIRVEHIIREDY 65
Query 70 LERAMEILVTVCELLISRLSYLATERT 96
+ A EIL C+LL++R + +T
Sbjct 66 VVEAFEILEMYCDLLLARFGLIEQMKT 92
> YNL265c
Length=298
Score = 35.8 bits (81), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 37/72 (51%), Gaps = 0/72 (0%)
Query 20 LKMAISRARMCTNRLQNSIRLQRKEIAGFLREGREEGARLKGEHLLREYRLERAMEILVT 79
LKM I R R + Q + R+++A L +E+ A + E L+ + +EIL
Sbjct 16 LKMCIQRLRYAQEKQQAIAKQSRRQVAQLLLTNKEQKAHYRVETLIHDDIHIELLEILEL 75
Query 80 VCELLISRLSYL 91
CELL++R+ +
Sbjct 76 YCELLLARVQVI 87
> At4g32350
Length=730
Score = 32.0 bits (71), Expect = 0.28, Method: Composition-based stats.
Identities = 18/89 (20%), Positives = 41/89 (46%), Gaps = 0/89 (0%)
Query 20 LKMAISRARMCTNRLQNSIRLQRKEIAGFLREGREEGARLKGEHLLREYRLERAMEILVT 79
+K+ +R + + +I+ ++++A + G + A + LL E R +++ +
Sbjct 16 IKLTKNRIDVLRRKRNATIKFLKRDLADLIINGHDYNAFSRAGGLLDELRYLWSLDFVEQ 75
Query 80 VCELLISRLSYLATERTCPPDLDSPIHTL 108
C+ + +LS + CP D I +L
Sbjct 76 TCDFVYKQLSTMQKTPECPEDCREAISSL 104
> Hs21553341
Length=6885
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 24/84 (28%), Positives = 41/84 (48%), Gaps = 10/84 (11%)
Query 18 RSLKMAISRARMCTN--RLQNSIRLQRKEIAGFLREGREEGARLKGEHLLRE----YRLE 71
+ LK+ I + ++ N +L+ I ++K +R GR +G + E E Y+L
Sbjct 938 QQLKIDIEKGKLSDNILKLEKQINKEKK----LIRRGRTKGLIKEHEACFSEEGCLYQLN 993
Query 72 RAMEILVTVCELLISRLSYLATER 95
ME+L +CE L S+ S +R
Sbjct 994 HHMEVLRELCEELPSQKSQQEVKR 1017
> At3g20350
Length=673
Score = 28.9 bits (63), Expect = 2.1, Method: Composition-based stats.
Identities = 17/56 (30%), Positives = 31/56 (55%), Gaps = 4/56 (7%)
Query 19 SLKMAISRARMCTNRLQNSIRLQRKEIAGFLREGREEGARLKGEHLLREYRLERAM 74
S+++ + AR C L++ R Q+K++ FL++ EE A + RE+ RA+
Sbjct 218 SIELKLQEARACIKDLESEKRSQKKKLEQFLKKVSEERAAWRS----REHEKVRAI 269
> 7296003
Length=1189
Score = 27.7 bits (60), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 17/40 (42%), Positives = 23/40 (57%), Gaps = 2/40 (5%)
Query 65 LREYRLERAMEILVTVCELLISRLSYLATERTCPPDLDSP 104
L+EY L+ +I V E +S L + T TCPPDL +P
Sbjct 852 LQEYTLDNRGDIGAWVREAAMSSLYEIVT--TCPPDLLAP 889
> At4g08450
Length=1234
Score = 27.3 bits (59), Expect = 6.3, Method: Composition-based stats.
Identities = 13/41 (31%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 60 KGEHLLREYRLERAMEILVTVCELLISRLSYLATERTCPPD 100
K +HLLR Y ++ E+L+ +L I A + PP+
Sbjct 327 KDKHLLRAYGIDHIYEVLLPSKDLAIKMFCRSAFRKDSPPN 367
Lambda K H
0.325 0.137 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160781780
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40