bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1255_orf1
Length=346
Score E
Sequences producing significant alignments: (Bits) Value
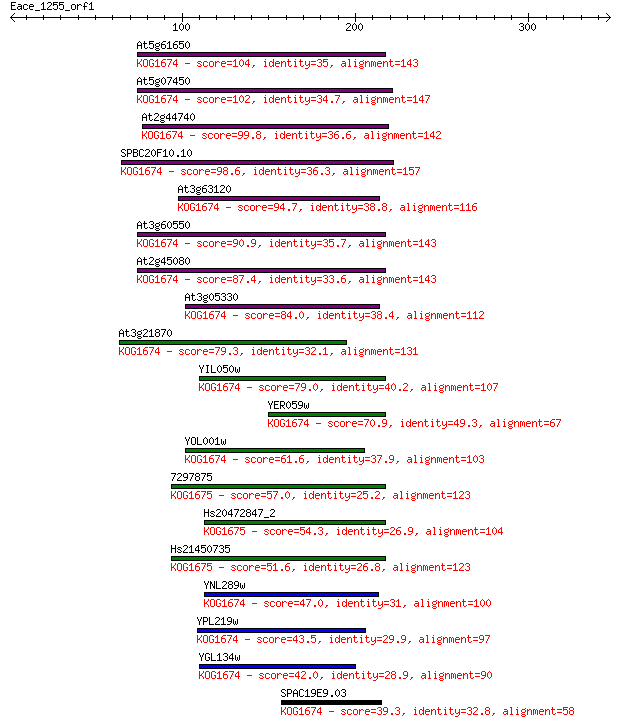
At5g61650 104 3e-22
At5g07450 102 9e-22
At2g44740 99.8 8e-21
SPBC20F10.10 98.6 2e-20
At3g63120 94.7 2e-19
At3g60550 90.9 4e-18
At2g45080 87.4 4e-17
At3g05330 84.0 4e-16
At3g21870 79.3 1e-14
YIL050w 79.0 1e-14
YER059w 70.9 4e-12
YOL001w 61.6 3e-09
7297875 57.0 6e-08
Hs20472847_2 54.3 4e-07
Hs21450735 51.6 2e-06
YNL289w 47.0 7e-05
YPL219w 43.5 6e-04
YGL134w 42.0 0.002
SPAC19E9.03 39.3 0.014
> At5g61650
Length=219
Score = 104 bits (259), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 50/146 (34%), Positives = 87/146 (59%), Gaps = 6/146 (4%)
Query 74 PLFVEKMSLLMEQLATCGDEPCRR--PSVATRFDSICPPATSIAAYIPRLYRHFRCSEEV 131
P + MS L+++++ D ++ PS F + P+ SI +Y+ R++ + CS
Sbjct 23 PSVLTAMSYLLQRVSETNDNLSQKQKPS---SFTGVTKPSISIRSYLERIFEYANCSYSC 79
Query 132 FVFALIYLDRVIRAN-HIKINALNIHRLVLAASVVGVKFVEDVRYSNRYYARVGGVGLTE 190
++ A IYLDR ++ + IN+ N+HRL++ + +V KF++D+ Y+N YYA+VGG+ E
Sbjct 80 YIVAYIYLDRFVKKQPFLPINSFNVHRLIITSVLVSAKFMDDLSYNNEYYAKVGGISREE 139
Query 191 LNRLELAFLKLVKFDLTVSKEEYAVY 216
+N LEL FL + F+L V+ + Y
Sbjct 140 MNMLELDFLFGIGFELNVTVSTFNNY 165
> At5g07450
Length=216
Score = 102 bits (255), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 51/151 (33%), Positives = 93/151 (61%), Gaps = 4/151 (2%)
Query 74 PLFVEKMSLLMEQLATCGDE---PCRRPSVATRFDSICPPATSIAAYIPRLYRHFRCSEE 130
P + MS L+++++ D+ P R + F+++ P+ SI +Y+ R++++ CS+
Sbjct 21 PNVITAMSSLLQRVSETNDDLSRPFREHKRISAFNAVTKPSISIRSYMERIFKYADCSDS 80
Query 131 VFVFALIYLDRVIRAN-HIKINALNIHRLVLAASVVGVKFVEDVRYSNRYYARVGGVGLT 189
++ A IYLDR I+ + I++ N+HRL++ + +V KF++D+ Y+N +YA+VGG+
Sbjct 81 CYIVAYIYLDRFIQKQPLLPIDSSNVHRLIITSVLVSAKFMDDLCYNNAFYAKVGGITTE 140
Query 190 ELNRLELAFLKLVKFDLTVSKEEYAVYRSTV 220
E+N LEL FL + F L V+ Y Y S++
Sbjct 141 EMNLLELDFLFGIGFQLNVTISTYNDYCSSL 171
> At2g44740
Length=202
Score = 99.8 bits (247), Expect = 8e-21, Method: Compositional matrix adjust.
Identities = 52/146 (35%), Positives = 86/146 (58%), Gaps = 4/146 (2%)
Query 77 VEKMSLLMEQLATCGDEPCRRPSVATR---FDSICPPATSIAAYIPRLYRHFRCSEEVFV 133
+ +S L+E++A D R + + R F + P +I +Y+ R++++ CS FV
Sbjct 14 IAFLSSLLERVAESNDLTRRVATQSQRVSVFHGLSRPTITIQSYLERIFKYANCSPSCFV 73
Query 134 FALIYLDRVI-RANHIKINALNIHRLVLAASVVGVKFVEDVRYSNRYYARVGGVGLTELN 192
A +YLDR R + IN+ N+HRL++ + +V KF++D+ Y+N YYA+VGG+ E+N
Sbjct 74 VAYVYLDRFTHRQPSLPINSFNVHRLLITSVMVAAKFLDDLYYNNAYYAKVGGISTKEMN 133
Query 193 RLELAFLKLVKFDLTVSKEEYAVYRS 218
LEL FL + F+L V+ + Y S
Sbjct 134 FLELDFLFGLGFELNVTPNTFNAYFS 159
> SPBC20F10.10
Length=243
Score = 98.6 bits (244), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 57/165 (34%), Positives = 90/165 (54%), Gaps = 8/165 (4%)
Query 65 GEEGVLPAGPLFVEKMSLLME--QLATCGDEPCRRPSVATR----FDSICPPATSIAAYI 118
+E +L +F+ +++ L + Q AT D+ P+ F + P+ SI AY+
Sbjct 27 NQEKLLEMISVFLSRLTRLNDSKQEATESDQIPLSPTSLKNPCLIFSAKNVPSISIQAYL 86
Query 119 PRLYRHFRCSEEVFVFALIYLDRVIRANH--IKINALNIHRLVLAASVVGVKFVEDVRYS 176
R+ ++ + +VF+ LIYLDR++ H + IN+ NIHR ++A KF DV Y+
Sbjct 87 TRILKYCPATNDVFLSVLIYLDRIVHHFHFTVFINSFNIHRFLIAGFTAASKFFSDVFYT 146
Query 177 NRYYARVGGVGLTELNRLELAFLKLVKFDLTVSKEEYAVYRSTVL 221
N YA+VGG+ L ELN LEL+F F+L +S E+ Y +L
Sbjct 147 NSRYAKVGGIPLHELNHLELSFFVFNDFNLFISLEDLQAYGDLLL 191
> At3g63120
Length=221
Score = 94.7 bits (234), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 45/116 (38%), Positives = 65/116 (56%), Gaps = 0/116 (0%)
Query 98 PSVATRFDSICPPATSIAAYIPRLYRHFRCSEEVFVFALIYLDRVIRANHIKINALNIHR 157
P T FD PP SIA Y+ R++++ CS FV A IY+D + + LN+HR
Sbjct 59 PDSVTVFDGRSPPEISIAHYLDRIFKYSCCSPSCFVIAHIYIDHFLHKTRALLKPLNVHR 118
Query 158 LVLAASVVGVKFVEDVRYSNRYYARVGGVGLTELNRLELAFLKLVKFDLTVSKEEY 213
L++ ++ K +D ++N YYARVGGV ELNRLE+ L + F L V + +
Sbjct 119 LIITTVMLAAKVFDDRYFNNAYYARVGGVTTRELNRLEMELLFTLDFKLQVDPQTF 174
> At3g60550
Length=230
Score = 90.9 bits (224), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 51/151 (33%), Positives = 85/151 (56%), Gaps = 8/151 (5%)
Query 74 PLFVEKMSLLMEQLATCGDEPCRR--PSVATR-----FDSICPPATSIAAYIPRLYRHFR 126
PL + +S L+++ T + RR PS FD P +I +Y+ R++R+ +
Sbjct 30 PLVISVLSSLIDRTLTRNERISRRALPSSGAGGKTQIFDCREIPDMTIQSYLGRIFRYTK 89
Query 127 CSEEVFVFALIYLDRVIRAN-HIKINALNIHRLVLAASVVGVKFVEDVRYSNRYYARVGG 185
V+V A +Y+DR + N +I+ N+HRL++ ++ K+VED+ Y N Y+A+VGG
Sbjct 90 AGPSVYVVAYVYIDRFCQTNPGFRISLTNVHRLLITTIMIASKYVEDLNYRNSYFAKVGG 149
Query 186 VGLTELNRLELAFLKLVKFDLTVSKEEYAVY 216
+ +LN+LEL FL L+ F L V+ + Y
Sbjct 150 LETEDLNKLELEFLFLMGFKLHVNVSVFESY 180
> At2g45080
Length=222
Score = 87.4 bits (215), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 48/147 (32%), Positives = 81/147 (55%), Gaps = 4/147 (2%)
Query 74 PLFVEKMSLLMEQLATCGDEPCRRPSVATR---FDSICPPATSIAAYIPRLYRHFRCSEE 130
PL + +S L+E+ + R + FD P +I +Y+ R++R+ +
Sbjct 28 PLVISVLSSLIERTLARNERISRSYGGFGKTRVFDCREIPDMTIQSYLERIFRYTKAGPS 87
Query 131 VFVFALIYLDRVIRANH-IKINALNIHRLVLAASVVGVKFVEDVRYSNRYYARVGGVGLT 189
V+V A +Y+DR + N +I+ N+HRL++ ++ K+VED+ Y N Y+A+VGG+
Sbjct 88 VYVVAYVYIDRFCQNNQGFRISLTNVHRLLITTIMIASKYVEDMNYKNSYFAKVGGLETE 147
Query 190 ELNRLELAFLKLVKFDLTVSKEEYAVY 216
+LN LEL FL L+ F L V+ + Y
Sbjct 148 DLNNLELEFLFLMGFKLHVNVSVFESY 174
> At3g05330
Length=588
Score = 84.0 bits (206), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 43/117 (36%), Positives = 68/117 (58%), Gaps = 5/117 (4%)
Query 102 TRFDSICPPATSIAAYIPRLYRHFRCSEEVFVFALIYLDRVIR-----ANHIKINALNIH 156
T F P+ SI Y R++R+ +CS FV A Y+ R ++ + ++ +LN+H
Sbjct 439 TMFHGSKAPSLSIYRYTERIHRYAQCSPVCFVAAFAYILRYLQRPEATSTARRLTSLNVH 498
Query 157 RLVLAASVVGVKFVEDVRYSNRYYARVGGVGLTELNRLELAFLKLVKFDLTVSKEEY 213
RL++ + +V KF+E Y+N YYA++GGV E+NRLE FL V F L ++ E +
Sbjct 499 RLLITSLLVAAKFLERQCYNNAYYAKIGGVSTEEMNRLERTFLVDVDFRLYITTETF 555
> At3g21870
Length=210
Score = 79.3 bits (194), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 42/136 (30%), Positives = 75/136 (55%), Gaps = 5/136 (3%)
Query 64 GGEEGVLPAGPLFVEKMSLLMEQLATCGDEPCRRPSVATR----FDSICPPATSIAAYIP 119
G E A P + +S +ME+L + ++ + F + P+ SIA Y+
Sbjct 15 GPESATEAATPRVLTIISHVMEKLVARNEWLAKQTKGFGKSLEAFHGVRAPSISIAKYLE 74
Query 120 RLYRHFRCSEEVFVFALIYLDRVIRAN-HIKINALNIHRLVLAASVVGVKFVEDVRYSNR 178
R+Y++ +CS FV +Y+DR+ + + +LN+HRL++ ++ K ++DV Y+N
Sbjct 75 RIYKYTKCSPACFVVGYVYIDRLAHKHPGSLVVSLNVHRLLVTCVMIAAKILDDVHYNNE 134
Query 179 YYARVGGVGLTELNRL 194
+YARVGGV +LN++
Sbjct 135 FYARVGGVSNADLNKM 150
> YIL050w
Length=285
Score = 79.0 bits (193), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 43/120 (35%), Positives = 64/120 (53%), Gaps = 14/120 (11%)
Query 110 PATSIAAYIPRLYRHFRCSEEVFVFALIYLDRVIRANH-------------IKINALNIH 156
P ++ Y+ R+ ++ + ++F+ L+Y DR I N+ +++ NIH
Sbjct 156 PEIAVVQYLERIQKYCPTTNDIFLSLLVYFDR-ISKNYGHSSERNGCAKQLFVMDSGNIH 214
Query 157 RLVLAASVVGVKFVEDVRYSNRYYARVGGVGLTELNRLELAFLKLVKFDLTVSKEEYAVY 216
RL++ + KF+ D YSN YA+VGG+ L ELN LEL FL L F L VS EE Y
Sbjct 215 RLLITGVTICTKFLSDFFYSNSRYAKVGGISLQELNHLELQFLILCDFKLLVSVEEMQKY 274
> YER059w
Length=420
Score = 70.9 bits (172), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 33/67 (49%), Positives = 43/67 (64%), Gaps = 0/67 (0%)
Query 150 INALNIHRLVLAASVVGVKFVEDVRYSNRYYARVGGVGLTELNRLELAFLKLVKFDLTVS 209
+++ NIHRL++A V KF+ D YSN Y+RVGG+ L ELN LEL FL L F+L +S
Sbjct 332 MDSHNIHRLIIAGITVSTKFLSDFFYSNSRYSRVGGISLQELNHLELQFLVLCDFELLIS 391
Query 210 KEEYAVY 216
E Y
Sbjct 392 VNELQRY 398
> YOL001w
Length=293
Score = 61.6 bits (148), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 39/104 (37%), Positives = 54/104 (51%), Gaps = 1/104 (0%)
Query 102 TRFDSICPPATSIAAYIPRLYRHFRCSEEVFVFALIYLDRVIRAN-HIKINALNIHRLVL 160
TR+ S PP SI Y RL + V + +L Y+D + +N+L HR +L
Sbjct 65 TRYHSKIPPNISIFNYFIRLTKFSSLEHCVLMTSLYYIDLLQTVYPDFTLNSLTAHRFLL 124
Query 161 AASVVGVKFVEDVRYSNRYYARVGGVGLTELNRLELAFLKLVKF 204
A+ V K + D +N +YA+VGGV ELN LE FLK V +
Sbjct 125 TATTVATKGLCDSFSTNAHYAKVGGVRCHELNILENDFLKRVNY 168
> 7297875
Length=406
Score = 57.0 bits (136), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 31/124 (25%), Positives = 60/124 (48%), Gaps = 1/124 (0%)
Query 94 PCRRPSVATRFDSICPPATSIAAYIPRLYRHFRCSEEVFVFALIYLDRVIRANHIKINAL 153
P V +D+ P I ++ L+ + + E + L+YL+R++ + +
Sbjct 213 PLTHDQVPDNYDTHNPEHRQIYKFVRTLFNAAQLTAECAIITLVYLERLLTYAELDVGPC 272
Query 154 NIHRLVLAASVVGVKFVEDVRYSNRYYARV-GGVGLTELNRLELAFLKLVKFDLTVSKEE 212
N R+VL A ++ K +D N Y ++ + + ++N LE FL+L++F++ V
Sbjct 273 NWKRMVLGAILLASKVWDDQAVWNVDYCQILKDITVEDMNELERQFLELLQFNINVPSSV 332
Query 213 YAVY 216
YA Y
Sbjct 333 YAKY 336
> Hs20472847_2
Length=301
Score = 54.3 bits (129), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 28/105 (26%), Positives = 58/105 (55%), Gaps = 4/105 (3%)
Query 113 SIAAYIPRLYRHFRCSEEVFVFALIYLDRVIRANHIKINALNIHRLVLAASVVGVKFVED 172
++ +++ L++ R + E FA+IY++R++ I I N R+VL A ++ K D
Sbjct 135 TLKSFVRTLFKAMRLTAE---FAIIYIERLVSYADIDICPTNWKRIVLGAILLASKVWSD 191
Query 173 VRYSNRYYARV-GGVGLTELNRLELAFLKLVKFDLTVSKEEYAVY 216
+ N Y ++ + + E+N LE FLKL+ ++ +++ Y+ +
Sbjct 192 MAVWNEDYCKLFKNITVEEMNELERQFLKLINYNNSITNSVYSRF 236
> Hs21450735
Length=341
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/124 (26%), Positives = 60/124 (48%), Gaps = 1/124 (0%)
Query 94 PCRRPSVATRFDSICPPATSIAAYIPRLYRHFRCSEEVFVFALIYLDRVIRANHIKINAL 153
P + V +D P I ++ L+ + + E + L+YL+R++ I I
Sbjct 151 PLSKSEVPPDYDKHNPEQKQIYRFVRTLFSAAQLTAECAIVTLVYLERLLTYAEIDICPA 210
Query 154 NIHRLVLAASVVGVKFVEDVRYSNRYYARV-GGVGLTELNRLELAFLKLVKFDLTVSKEE 212
N R+VL A ++ K +D N Y ++ + + ++N LE FL+L++F++ V
Sbjct 211 NWKRIVLGAILLASKVWDDQAVWNVDYCQILKDITVEDMNELERQFLELLQFNINVPSSV 270
Query 213 YAVY 216
YA Y
Sbjct 271 YAKY 274
> YNL289w
Length=279
Score = 47.0 bits (110), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 31/104 (29%), Positives = 52/104 (50%), Gaps = 4/104 (3%)
Query 113 SIAAYIPRLYRHFRCSEEVFVFALIYLDRVIRANHIKINAL--NIHRLVLAASVVGVKFV 170
S+ +I RL R+ + A YL+++ R L IHR+ LA ++ KF
Sbjct 54 SLMTFITRLVRYTNVYTPTLLTAACYLNKLKRILPRDATGLPSTIHRIFLACLILSAKFH 113
Query 171 EDVRYSNRYYARV--GGVGLTELNRLELAFLKLVKFDLTVSKEE 212
D N+++AR G L ++N +E L+L+ +DL V+ E+
Sbjct 114 NDSSPLNKHWARYTDGLFTLEDINLMERQLLQLLNWDLRVNTED 157
> YPL219w
Length=492
Score = 43.5 bits (101), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 29/106 (27%), Positives = 50/106 (47%), Gaps = 9/106 (8%)
Query 109 PPATSIAAYIPRLYRHFRCSEEVFVFA-----LIYLDRVIRANHIKINAL----NIHRLV 159
PP+ S +I R+ V++ A L++L R IK+ A HR++
Sbjct 361 PPSLSFRDFIDRIQNKCMFGAVVYLGATYLLQLVFLTRDEMDGPIKLKAKLQEDQAHRII 420
Query 160 LAASVVGVKFVEDVRYSNRYYARVGGVGLTELNRLELAFLKLVKFD 205
++ + K +ED +S Y +V G+ L +LE++F+ V FD
Sbjct 421 ISTIRIATKLLEDFVHSQNYICKVFGISKRLLTKLEISFMASVNFD 466
> YGL134w
Length=433
Score = 42.0 bits (97), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/98 (26%), Positives = 49/98 (50%), Gaps = 8/98 (8%)
Query 110 PATSIAAYIPRLYRHFRCSEEVFVFALIYLDRVI---RANHIKINALN-----IHRLVLA 161
P S A ++ R+ V++ A +D + N+I LN +HR+++A
Sbjct 306 PTLSSADFLKRIQDKCEYQPTVYLVATFLIDTLFLTRDGNNILQLKLNLQEKEVHRMIIA 365
Query 162 ASVVGVKFVEDVRYSNRYYARVGGVGLTELNRLELAFL 199
A + K +ED +S+ Y+++V G+ L +LE++ L
Sbjct 366 AVRLSTKLLEDFVHSHEYFSKVCGISKRLLTKLEVSLL 403
> SPAC19E9.03
Length=166
Score = 39.3 bits (90), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 33/58 (56%), Gaps = 0/58 (0%)
Query 157 RLVLAASVVGVKFVEDVRYSNRYYARVGGVGLTELNRLELAFLKLVKFDLTVSKEEYA 214
RL L + KF++D +SNR ++R+ G+ + +L LE F + + + L V K +A
Sbjct 85 RLFLVCLMAATKFLQDRSFSNRAWSRLSGLPVDKLLVLEYMFYQCIDYRLVVPKHIFA 142
Lambda K H
0.319 0.131 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8340552822
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40