bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
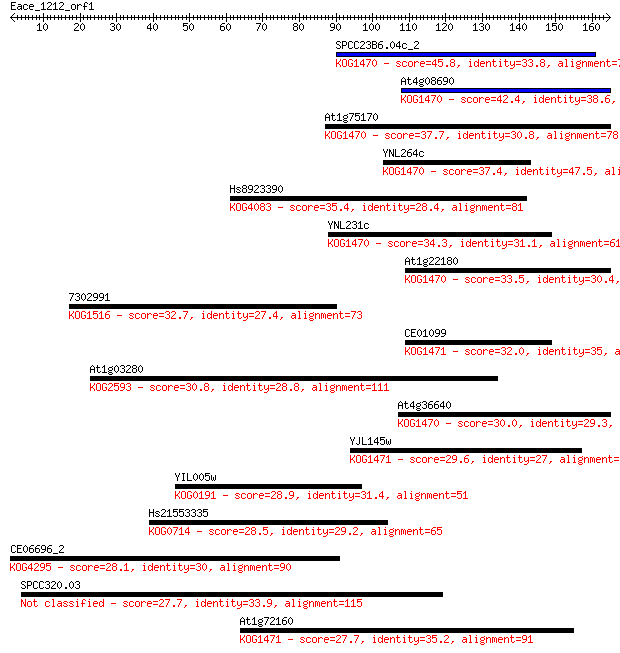
Query= Eace_1212_orf1
Length=164
Score E
Sequences producing significant alignments: (Bits) Value
SPCC23B6.04c_2 45.8 4e-05
At4g08690 42.4 4e-04
At1g75170 37.7 0.011
YNL264c 37.4 0.012
Hs8923390 35.4 0.054
YNL231c 34.3 0.12
At1g22180 33.5 0.17
7302991 32.7 0.33
CE01099 32.0 0.63
At1g03280 30.8 1.3
At4g36640 30.0 2.2
YJL145w 29.6 2.8
YIL005w 28.9 5.4
Hs21553335 28.5 5.8
CE06696_2 28.1 8.4
SPCC320.03 27.7 9.2
At1g72160 27.7 9.8
> SPCC23B6.04c_2
Length=442
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 44/71 (61%), Gaps = 3/71 (4%)
Query 90 SSNTTKKEVEELTWEEVLWLSNDLILWRYLRSYNWEEAVAQQQLLQTIIWRRERKPHHIH 149
+SN++KK +L E LWL+ + IL RYLR+ W + A+++++ T++WRR +++
Sbjct 50 ASNSSKKT--DLIELERLWLTRECIL-RYLRATKWHVSNAKKRIVDTLVWRRHFGVNNMD 106
Query 150 PKDVMTTAARG 160
P ++ A G
Sbjct 107 PDEIQEENATG 117
> At4g08690
Length=301
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 22/57 (38%), Positives = 33/57 (57%), Gaps = 1/57 (1%)
Query 108 WLSNDLILWRYLRSYNWEEAVAQQQLLQTIIWRRERKPHHIHPKDVMTTAARGSVYR 164
+ S+D +L RYLR+ NW A + L +T+ WR + KP I ++V A G +YR
Sbjct 40 FCSDDAVL-RYLRARNWHVKKATKMLKETLKWRVQYKPEEICWEEVAGEAETGKIYR 95
> At1g75170
Length=296
Score = 37.7 bits (86), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 36/78 (46%), Gaps = 1/78 (1%)
Query 87 RSSSSNTTKKEVEELTWEEVLWLSNDLILWRYLRSYNWEEAVAQQQLLQTIIWRRERKPH 146
R + K + +L+ L+ S D L RYL + NW A++ L +T+ WR KP
Sbjct 20 REAKMKELKTLIGQLSGRNSLYCS-DACLKRYLEARNWNVGKAKKMLEETLKWRSSFKPE 78
Query 147 HIHPKDVMTTAARGSVYR 164
I +V G VY+
Sbjct 79 EIRWNEVSGEGETGKVYK 96
> YNL264c
Length=350
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 19/40 (47%), Positives = 26/40 (65%), Gaps = 2/40 (5%)
Query 103 WEEVLWLSNDLILWRYLRSYNWEEAVAQQQLLQTIIWRRE 142
WE+ WLS + L RYLR+ W A A + L +T++WRRE
Sbjct 85 WEK-FWLSRECFL-RYLRANKWNTANAIKGLTKTLVWRRE 122
> Hs8923390
Length=227
Score = 35.4 bits (80), Expect = 0.054, Method: Compositional matrix adjust.
Identities = 23/82 (28%), Positives = 39/82 (47%), Gaps = 1/82 (1%)
Query 61 SFPIVDELREEAPAAKEKSWLSGMFSRSSSSNTTKKEV-EELTWEEVLWLSNDLILWRYL 119
S ++D ++E +P+ + SG + S S K+ V EEL E+ S D +
Sbjct 29 SENVIDRMKESSPSGSKSQRYSGAYGASVSDEELKRRVAEELALEQAKKESEDQKRLKQA 88
Query 120 RSYNWEEAVAQQQLLQTIIWRR 141
+ + E A A +QL + I+ R
Sbjct 89 KELDRERAAANEQLTRAILRER 110
> YNL231c
Length=351
Score = 34.3 bits (77), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 32/61 (52%), Gaps = 1/61 (1%)
Query 88 SSSSNTTKKEVEELTWEEVLWLSNDLILWRYLRSYNWEEAVAQQQLLQTIIWRRERKPHH 147
+S N ++ +++ L EE WL+ + L RYLR+ W ++ T+ WRRE H
Sbjct 64 TSEKNKSEDDLKPLEEEEKAWLTRECFL-RYLRATKWVLKDCIDRITMTLAWRREFGISH 122
Query 148 I 148
+
Sbjct 123 L 123
> At1g22180
Length=314
Score = 33.5 bits (75), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 28/56 (50%), Gaps = 0/56 (0%)
Query 109 LSNDLILWRYLRSYNWEEAVAQQQLLQTIIWRRERKPHHIHPKDVMTTAARGSVYR 164
+D + RYL + N A + L +T+ WR + KP I +++ A G +YR
Sbjct 43 FCSDAAITRYLAARNGHVKKATKMLKETLKWRAQYKPEEIRWEEIAREAETGKIYR 98
> 7302991
Length=803
Score = 32.7 bits (73), Expect = 0.33, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 33/74 (44%), Gaps = 3/74 (4%)
Query 17 PQPLCTATGPPPSLGIFPP-IESVEGLNLNAEQLQKVRSLRAMVDSFPIVDELREEAPAA 75
P P C A L + PP + ++G NL Q ++ + + + P + +LR P
Sbjct 14 PGPFCAALATVDQLTVCPPSVGCLKGTNLQGYQSERFEAFMGIPYALPPIGDLRFSNPKV 73
Query 76 KEKSWLSGMFSRSS 89
K L GM+ S+
Sbjct 74 MPK--LLGMYDASA 85
> CE01099
Length=743
Score = 32.0 bits (71), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 24/40 (60%), Gaps = 0/40 (0%)
Query 109 LSNDLILWRYLRSYNWEEAVAQQQLLQTIIWRRERKPHHI 148
L ND L R+LR+ +++ A A+ + +IIWR++ I
Sbjct 298 LPNDAHLLRFLRARDFDVAKAKDMVHASIIWRKQHNVDKI 337
> At1g03280
Length=506
Score = 30.8 bits (68), Expect = 1.3, Method: Composition-based stats.
Identities = 32/113 (28%), Positives = 53/113 (46%), Gaps = 17/113 (15%)
Query 23 ATGPPPSLGIFPPIESVEGLNLNAEQLQKVRSLRAMVDSFPIVDELREEAPAAKEKSWLS 82
+ G SL + PP EG+NL EQ ++R A VD A AA + S
Sbjct 323 SKGGDSSLKVLPPWMIKEGMNLTEEQRGEMRQ-EAKVDG---------GAGAAAKLS--- 369
Query 83 GMFSRSSSSNTTKKEVEELTWEEVL--WLSNDLILWRYLRSYNWEEAVAQQQL 133
+S+ N +K+++ W ++L +L + YL++Y + E + QQ+L
Sbjct 370 -DDKKSAIGNGDEKDLKACFWGQILYIYLDSHSSFDEYLKAY-YAELMKQQEL 420
> At4g36640
Length=294
Score = 30.0 bits (66), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 26/58 (44%), Gaps = 0/58 (0%)
Query 107 LWLSNDLILWRYLRSYNWEEAVAQQQLLQTIIWRRERKPHHIHPKDVMTTAARGSVYR 164
L +D L R+L + NW+ A++ + +T+ WR KP I V G R
Sbjct 36 LVFCSDASLRRFLDARNWDVEKAKKMIQETLKWRSTYKPQEIRWNQVAHEGETGKASR 93
> YJL145w
Length=294
Score = 29.6 bits (65), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 17/63 (26%), Positives = 33/63 (52%), Gaps = 4/63 (6%)
Query 94 TKKEVEELTWEEVLWLSNDLILWRYLRSYNWEEAVAQQQLLQTIIWRRERKPHHIHPKDV 153
T++EV++ E++ D + ++ ++Y +E + Q L+ + WRRE P K+V
Sbjct 44 TQEEVDKYYDEKIA----DRLTYKLCKAYQFEYSTIVQNLIDILNWRREFNPLSCAYKEV 99
Query 154 MTT 156
T
Sbjct 100 HNT 102
> YIL005w
Length=701
Score = 28.9 bits (63), Expect = 5.4, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 27/51 (52%), Gaps = 2/51 (3%)
Query 46 AEQLQKVRSLRAMVDSFPIVDELREEAPAAKEKSWLSGMFSRSSSSNTTKK 96
A+ ++K++ +A + IVD++REE P +K L G S N +K
Sbjct 521 ADGIKKIKEKKAPANK--IVDKMREEIPHMDQKKLLLGYLDISKEKNFFRK 569
> Hs21553335
Length=309
Score = 28.5 bits (62), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 19/69 (27%), Positives = 35/69 (50%), Gaps = 4/69 (5%)
Query 39 VEGLNLNAEQLQKVRSLRAMVD----SFPIVDELREEAPAAKEKSWLSGMFSRSSSSNTT 94
V G N+N +++ + R D +F +V+ + E AKE SW + F+ S ++ +
Sbjct 194 VNGRNINTKKIIESDQEREAEDNGELTFFLVNSVANEEGFAKECSWRTQSFNNYSPNSHS 253
Query 95 KKEVEELTW 103
K V + T+
Sbjct 254 SKHVSQYTF 262
> CE06696_2
Length=185
Score = 28.1 bits (61), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 27/94 (28%), Positives = 39/94 (41%), Gaps = 16/94 (17%)
Query 1 CPSPSSSPPPGTSSTSPQPLCTATGPPP----SLGIFPPIESVEGLNLNAEQLQKVRSLR 56
CP PP ++T+P P T+ G P S+G F I E L N Q
Sbjct 82 CPEGPDGPPGFKTTTTPGPKLTSDGCPKGYICSMGAFFGICCEENL-TNRYQA------- 133
Query 57 AMVDSFPIVDELREEAPAAKEKSWLSGMFSRSSS 90
+F + + ++EA K+ W S M +S S
Sbjct 134 ----AFKPLCKNKKEALQTKQDDWSSAMLGKSCS 163
> SPCC320.03
Length=867
Score = 27.7 bits (60), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 39/124 (31%), Positives = 55/124 (44%), Gaps = 19/124 (15%)
Query 4 PSSSPPPGTSSTSPQP--------LCTATGPPPSLGIFPP-IESVEGLNLNAEQLQKVRS 54
PS+SP S T+ P L + P PSLG+ P + ++ LN N + +V S
Sbjct 353 PSNSPATLNSYTTSVPSGMSRHPMLMNPSTPEPSLGVNSPSLRPLQSLN-NVQNSYRVAS 411
Query 55 LRAMVDSFPIVDELREEAPAAKEKSWLSGMFSRSSSSNTTKKEVEELTWEEVLWLSNDLI 114
+A P LR A+ S S S +S T +EVE+L E V W +D
Sbjct 412 TQA-----PPPHPLRNYTSDAESISMRSK--STQASDAATFREVEQLYQENVEW--DDAA 462
Query 115 LWRY 118
+ RY
Sbjct 463 IDRY 466
> At1g72160
Length=490
Score = 27.7 bits (60), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 32/122 (26%), Positives = 56/122 (45%), Gaps = 31/122 (25%)
Query 64 IVDELREEAPAAKEKSWLS---GMFSRSSS-----SNTTKKEVEEL-------------- 101
+ DE ++ A ++KS + G F SS SN+ KK ++EL
Sbjct 85 VKDEASQKEVAEEKKSMIPQNLGSFKEESSKLSDLSNSEKKSLDELKHLVREALDNHQFT 144
Query 102 -TWEEV-LW-------LSNDLILWRYLRSYNWEEAVAQQQLLQTIIWRRERKPHHIHPKD 152
T EEV +W +D++L ++LR+ ++ + L TI WR+E K + +D
Sbjct 145 NTPEEVKIWGIPLLEDDRSDVVLLKFLRAREFKVKDSFAMLKNTIKWRKEFKIDELVEED 204
Query 153 VM 154
++
Sbjct 205 LV 206
Lambda K H
0.313 0.128 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2317343104
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40