bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1204_orf1
Length=201
Score E
Sequences producing significant alignments: (Bits) Value
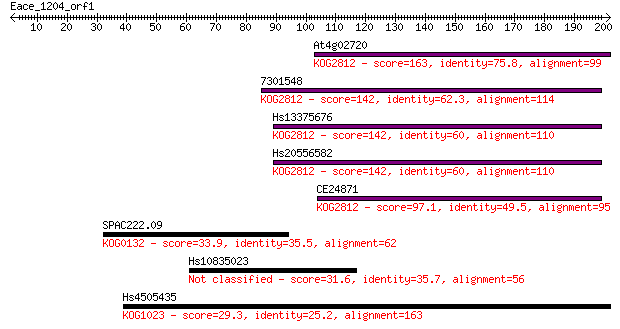
At4g02720 163 3e-40
7301548 142 4e-34
Hs13375676 142 5e-34
Hs20556582 142 6e-34
CE24871 97.1 2e-20
SPAC222.09 33.9 0.25
Hs10835023 31.6 1.1
Hs4505435 29.3 5.8
> At4g02720
Length=420
Score = 163 bits (412), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 75/99 (75%), Positives = 88/99 (88%), Gaps = 0/99 (0%)
Query 103 AQKNMDYGFALRPGEGEAIAQFVQEGKRIPRRGEVGLTAEEISNFESLGYVMSGSRHRRM 162
A+ ++ YG ALRPGEG+AIAQ+VQ+GKRIPRRGEVGL AEEI FE LGYVMSGSRH+RM
Sbjct 293 AEGHISYGGALRPGEGDAIAQYVQQGKRIPRRGEVGLNAEEIQKFEDLGYVMSGSRHQRM 352
Query 163 NAIRIRKENQVYSAEEQRALAMLNYEERAARETQLLYDL 201
NAIRIRKENQVYSAE++RALAM NYEE+A RE +++ DL
Sbjct 353 NAIRIRKENQVYSAEDKRALAMFNYEEKAKREAKVMSDL 391
> 7301548
Length=463
Score = 142 bits (359), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 71/114 (62%), Positives = 89/114 (78%), Gaps = 3/114 (2%)
Query 85 SSEDDMAGPRPMEVTNKLAQKNMDYGFALRPGEGEAIAQFVQEGKRIPRRGEVGLTAEEI 144
S D+ GP + L QK D+G AL PGEG A+A ++ EGKRIPRRGE+GLT++EI
Sbjct 336 SHNDEDVGP-SLRPGGSLNQK--DFGKALLPGEGAAMAAYIAEGKRIPRRGEIGLTSDEI 392
Query 145 SNFESLGYVMSGSRHRRMNAIRIRKENQVYSAEEQRALAMLNYEERAARETQLL 198
+NFES+GYVMSGSRHRRM A+RIRKENQ+YSA+E+RALAM + EER RE ++L
Sbjct 393 ANFESVGYVMSGSRHRRMEAVRIRKENQLYSADEKRALAMFSKEERQKRENKIL 446
> Hs13375676
Length=415
Score = 142 bits (357), Expect = 5e-34, Method: Compositional matrix adjust.
Identities = 66/110 (60%), Positives = 85/110 (77%), Gaps = 0/110 (0%)
Query 89 DMAGPRPMEVTNKLAQKNMDYGFALRPGEGEAIAQFVQEGKRIPRRGEVGLTAEEISNFE 148
D+ GP + K ++YG AL PGEG A+A++V+ GKRIPRRGE+GLT+EEI++FE
Sbjct 289 DLIGPEAPKTLTSQDDKPLNYGHALLPGEGAAMAEYVKAGKRIPRRGEIGLTSEEIASFE 348
Query 149 SLGYVMSGSRHRRMNAIRIRKENQVYSAEEQRALAMLNYEERAARETQLL 198
GYVMSGSRHRRM A+R+RKENQ+YSA+E+RALA N EER RE ++L
Sbjct 349 CSGYVMSGSRHRRMEAVRLRKENQIYSADEKRALASFNQEERRKRENKIL 398
> Hs20556582
Length=362
Score = 142 bits (357), Expect = 6e-34, Method: Compositional matrix adjust.
Identities = 66/110 (60%), Positives = 85/110 (77%), Gaps = 0/110 (0%)
Query 89 DMAGPRPMEVTNKLAQKNMDYGFALRPGEGEAIAQFVQEGKRIPRRGEVGLTAEEISNFE 148
D+ GP + +K + YG AL PGEG A+A++V+ GKRIPRRGE+GLT+EEI +FE
Sbjct 236 DLIGPEAPIIHTSQDEKPLKYGHALLPGEGAAMAEYVKAGKRIPRRGEIGLTSEEIGSFE 295
Query 149 SLGYVMSGSRHRRMNAIRIRKENQVYSAEEQRALAMLNYEERAARETQLL 198
GYVMSGSRHRRM A+R+RKENQ+YSA+E+RALA N EER RE+++L
Sbjct 296 CSGYVMSGSRHRRMEAVRLRKENQIYSADEKRALASFNQEERRKRESKIL 345
> CE24871
Length=504
Score = 97.1 bits (240), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 47/95 (49%), Positives = 68/95 (71%), Gaps = 0/95 (0%)
Query 104 QKNMDYGFALRPGEGEAIAQFVQEGKRIPRRGEVGLTAEEISNFESLGYVMSGSRHRRMN 163
+K +YG L GEG +A + G+RIPRRGE+GL++ EI+ +E +GYVMSGSRH+ M
Sbjct 396 EKPGNYGKDLLKGEGAGMAAYAARGERIPRRGEIGLSSGEIAEYEKVGYVMSGSRHKAME 455
Query 164 AIRIRKENQVYSAEEQRALAMLNYEERAARETQLL 198
A R+RKENQV++AEE+R L ++ E + +E +L
Sbjct 456 ATRLRKENQVFTAEEKRILNSVSVEAKKKKEEAVL 490
> SPAC222.09
Length=620
Score = 33.9 bits (76), Expect = 0.25, Method: Composition-based stats.
Identities = 22/66 (33%), Positives = 30/66 (45%), Gaps = 4/66 (6%)
Query 32 AAAAAATSPAAATGAAAADAAAPASRAATRPNVAVLEAAAAEMQH----KAEEEEGSSSE 87
AA + +T+P G+ + AAAP SR ++LEA AA Q A EE S+
Sbjct 163 AAPSQSTNPEGNGGSVGSQAAAPTSRPVENDAASILEALAAFAQKAPVPSAAEESVSTPP 222
Query 88 DDMAGP 93
P
Sbjct 223 QPAVAP 228
> Hs10835023
Length=2695
Score = 31.6 bits (70), Expect = 1.1, Method: Composition-based stats.
Identities = 20/61 (32%), Positives = 30/61 (49%), Gaps = 5/61 (8%)
Query 61 RPNVAVLEAAAAEMQHKAEEEEGSSSEDDMAGPRPME-----VTNKLAQKNMDYGFALRP 115
RP V A MQ + E E+G + ED A PR + + ++LA+ N + L+P
Sbjct 2062 RPKELVEVIKKAYMQGEVEFEDGENGEDGAASPRNVGHNIYILAHQLARHNKELQSMLKP 2121
Query 116 G 116
G
Sbjct 2122 G 2122
> Hs4505435
Length=1061
Score = 29.3 bits (64), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 41/174 (23%), Positives = 68/174 (39%), Gaps = 20/174 (11%)
Query 39 SPAAATGAAAADAAAPASRAATRPNVAVLEAAAAEMQHKAEEEEGSSSEDDMAGPRPMEV 98
+PA G AAAP R V +L A A + ++E ++ AGP ++
Sbjct 109 NPAVFLGPGCVYAAAPVGRFTAHWRVPLLTAGAPALGFGVKDEYALTTR---AGPSYAKL 165
Query 99 TNKLAQKNMDYG--------FALRPGEGEAIAQFVQEGKRIPRRGEVGLTAEEISNFESL 150
+ +A + G +A RPG+ E F+ EG + R + +T + + E
Sbjct 166 GDFVAALHRRLGWERQALMLYAYRPGDEEHCF-FLVEGLFMRVRDRLNITVDHLEFAED- 223
Query 151 GYVMSGSRHRRMNAIRIRKENQVY---SAEEQRALAMLNYEERAARETQLLYDL 201
S + R+ RK +Y S + R L +L E E + + L
Sbjct 224 ----DLSHYTRLLRTMPRKGRVIYICSSPDAFRTLMLLALEAGLCGEDYVFFHL 273
Lambda K H
0.318 0.128 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3515233148
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40