bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1189_orf2
Length=64
Score E
Sequences producing significant alignments: (Bits) Value
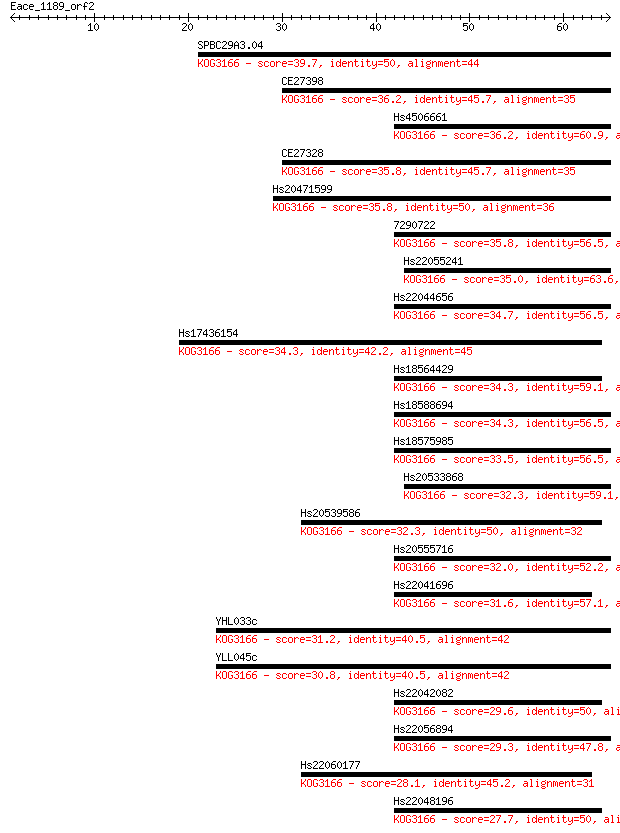
SPBC29A3.04 39.7 0.001
CE27398 36.2 0.016
Hs4506661 36.2 0.016
CE27328 35.8 0.017
Hs20471599 35.8 0.020
7290722 35.8 0.020
Hs22055241 35.0 0.030
Hs22044656 34.7 0.044
Hs17436154 34.3 0.052
Hs18564429 34.3 0.057
Hs18588694 34.3 0.061
Hs18575985 33.5 0.085
Hs20533868 32.3 0.20
Hs20539586 32.3 0.21
Hs20555716 32.0 0.32
Hs22041696 31.6 0.39
YHL033c 31.2 0.47
YLL045c 30.8 0.67
Hs22042082 29.6 1.4
Hs22056894 29.3 2.0
Hs22060177 28.1 3.7
Hs22048196 27.7 5.2
> SPBC29A3.04
Length=259
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/44 (50%), Positives = 29/44 (65%), Gaps = 0/44 (0%)
Query 21 SKKKLPSNPAGKNVKKTTKSLIFQKTPRNFRIGCNIQPRRELTR 64
SKK PS A KTTK+ +F PR+F IG +IQP+R+L+R
Sbjct 5 SKKVAPSPFAQPKAAKTTKNPLFVSRPRSFGIGQDIQPKRDLSR 48
> CE27398
Length=265
Score = 36.2 bits (82), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 16/35 (45%), Positives = 25/35 (71%), Gaps = 0/35 (0%)
Query 30 AGKNVKKTTKSLIFQKTPRNFRIGCNIQPRRELTR 64
A V+K K+ +F+K RNF IG +IQP++++TR
Sbjct 20 AQTQVQKEVKNPLFEKRARNFNIGQDIQPKKDVTR 54
> Hs4506661
Length=266
Score = 36.2 bits (82), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 14/23 (60%), Positives = 20/23 (86%), Gaps = 0/23 (0%)
Query 42 IFQKTPRNFRIGCNIQPRRELTR 64
+F+K P+NF IG +IQP+R+LTR
Sbjct 31 LFEKRPKNFGIGQDIQPKRDLTR 53
> CE27328
Length=234
Score = 35.8 bits (81), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 16/35 (45%), Positives = 25/35 (71%), Gaps = 0/35 (0%)
Query 30 AGKNVKKTTKSLIFQKTPRNFRIGCNIQPRRELTR 64
A V+K K+ +F+K RNF IG +IQP++++TR
Sbjct 123 AQTQVQKEVKNPLFEKRARNFNIGQDIQPKKDVTR 157
> Hs20471599
Length=259
Score = 35.8 bits (81), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 18/38 (47%), Positives = 25/38 (65%), Gaps = 2/38 (5%)
Query 29 PAGKNVKKTTKSL--IFQKTPRNFRIGCNIQPRRELTR 64
PAG ++ K + +F+K P NF IG +IQP+RELT
Sbjct 16 PAGMKKQEAKKVVNPLFEKRPENFGIGQDIQPKRELTH 53
> 7290722
Length=271
Score = 35.8 bits (81), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 13/23 (56%), Positives = 20/23 (86%), Gaps = 0/23 (0%)
Query 42 IFQKTPRNFRIGCNIQPRRELTR 64
+F+K P+NF IG N+QP+R+L+R
Sbjct 36 LFEKRPKNFGIGQNVQPKRDLSR 58
> Hs22055241
Length=260
Score = 35.0 bits (79), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 14/22 (63%), Positives = 19/22 (86%), Gaps = 0/22 (0%)
Query 43 FQKTPRNFRIGCNIQPRRELTR 64
F+K P+NF IG +IQP+R+LTR
Sbjct 32 FEKRPKNFVIGRDIQPKRDLTR 53
> Hs22044656
Length=178
Score = 34.7 bits (78), Expect = 0.044, Method: Compositional matrix adjust.
Identities = 13/23 (56%), Positives = 20/23 (86%), Gaps = 0/23 (0%)
Query 42 IFQKTPRNFRIGCNIQPRRELTR 64
+F++ P+NF IG +IQP+R+LTR
Sbjct 31 LFEERPKNFGIGQDIQPKRDLTR 53
> Hs17436154
Length=134
Score = 34.3 bits (77), Expect = 0.052, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 29/47 (61%), Gaps = 2/47 (4%)
Query 19 GASKKKLPSNPAGKNVKKTTKSL--IFQKTPRNFRIGCNIQPRRELT 63
G KK+ PA ++ K + +F+K P+NF IG +IQP+R+LT
Sbjct 4 GKKGKKVAPAPAVMTKQEAKKVVNPLFEKRPKNFGIGQDIQPKRDLT 50
> Hs18564429
Length=294
Score = 34.3 bits (77), Expect = 0.057, Method: Compositional matrix adjust.
Identities = 13/22 (59%), Positives = 19/22 (86%), Gaps = 0/22 (0%)
Query 42 IFQKTPRNFRIGCNIQPRRELT 63
+F+K P+NF IG +IQP+R+LT
Sbjct 61 LFEKRPKNFDIGQDIQPKRDLT 82
> Hs18588694
Length=266
Score = 34.3 bits (77), Expect = 0.061, Method: Compositional matrix adjust.
Identities = 13/23 (56%), Positives = 19/23 (82%), Gaps = 0/23 (0%)
Query 42 IFQKTPRNFRIGCNIQPRRELTR 64
+ +K P+NF IG +IQP+R+LTR
Sbjct 31 LLEKRPKNFGIGQDIQPKRDLTR 53
> Hs18575985
Length=145
Score = 33.5 bits (75), Expect = 0.085, Method: Compositional matrix adjust.
Identities = 13/23 (56%), Positives = 18/23 (78%), Gaps = 0/23 (0%)
Query 42 IFQKTPRNFRIGCNIQPRRELTR 64
+F+K P+NF IG +IQP+R TR
Sbjct 13 LFEKRPKNFGIGQDIQPKRHFTR 35
> Hs20533868
Length=229
Score = 32.3 bits (72), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 13/22 (59%), Positives = 18/22 (81%), Gaps = 0/22 (0%)
Query 43 FQKTPRNFRIGCNIQPRRELTR 64
F+K P+NF IG +IQP+ +LTR
Sbjct 14 FEKRPKNFGIGQDIQPKSDLTR 35
> Hs20539586
Length=199
Score = 32.3 bits (72), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 16/32 (50%), Positives = 23/32 (71%), Gaps = 1/32 (3%)
Query 32 KNVKKTTKSLIFQKTPRNFRIGCNIQPRRELT 63
+ VKK L+ +K P+NF IG +IQP+R+LT
Sbjct 3 QEVKKVVNPLL-EKRPKNFGIGQDIQPKRDLT 33
> Hs20555716
Length=192
Score = 32.0 bits (71), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 12/23 (52%), Positives = 18/23 (78%), Gaps = 0/23 (0%)
Query 42 IFQKTPRNFRIGCNIQPRRELTR 64
+F+K P+NF G +IQP+R+LT
Sbjct 31 LFEKRPKNFGTGQDIQPKRDLTH 53
> Hs22041696
Length=113
Score = 31.6 bits (70), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 12/21 (57%), Positives = 18/21 (85%), Gaps = 0/21 (0%)
Query 42 IFQKTPRNFRIGCNIQPRREL 62
+F+K P+NF IG +IQP+R+L
Sbjct 31 LFEKRPKNFGIGQDIQPKRDL 51
> YHL033c
Length=256
Score = 31.2 bits (69), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 26/44 (59%), Gaps = 2/44 (4%)
Query 23 KKLPSNPAGKNVKKT--TKSLIFQKTPRNFRIGCNIQPRRELTR 64
KK+ P G K+ T++ + TP+NF IG +QP+R L+R
Sbjct 5 KKVAPAPFGAKSTKSNKTRNPLTHSTPKNFGIGQAVQPKRNLSR 48
> YLL045c
Length=256
Score = 30.8 bits (68), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 25/44 (56%), Gaps = 2/44 (4%)
Query 23 KKLPSNPAGKNVKKTTKSL--IFQKTPRNFRIGCNIQPRRELTR 64
KK+ P G K+ K+ + TP+NF IG +QP+R L+R
Sbjct 5 KKVAPAPFGAKSTKSNKAKNPLTHSTPKNFGIGQAVQPKRNLSR 48
> Hs22042082
Length=195
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 11/22 (50%), Positives = 17/22 (77%), Gaps = 0/22 (0%)
Query 42 IFQKTPRNFRIGCNIQPRRELT 63
+F+K P+NF +IQP+R+LT
Sbjct 30 LFEKRPKNFGTEQDIQPKRDLT 51
> Hs22056894
Length=192
Score = 29.3 bits (64), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 11/23 (47%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 42 IFQKTPRNFRIGCNIQPRRELTR 64
+F+K P+N G +IQP+R LT
Sbjct 31 LFEKRPKNLGTGQDIQPKRHLTH 53
> Hs22060177
Length=223
Score = 28.1 bits (61), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 21/31 (67%), Gaps = 1/31 (3%)
Query 32 KNVKKTTKSLIFQKTPRNFRIGCNIQPRREL 62
++ KK SL F+K P+NF G +IQP++ L
Sbjct 22 QDAKKVVNSL-FEKRPKNFGTGEDIQPQKLL 51
> Hs22048196
Length=220
Score = 27.7 bits (60), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 11/22 (50%), Positives = 16/22 (72%), Gaps = 0/22 (0%)
Query 42 IFQKTPRNFRIGCNIQPRRELT 63
+F+K P+N IG +I+P ELT
Sbjct 34 LFEKKPKNSGIGQDIRPESELT 55
Lambda K H
0.319 0.130 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1169706042
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40