bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1182_orf1
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
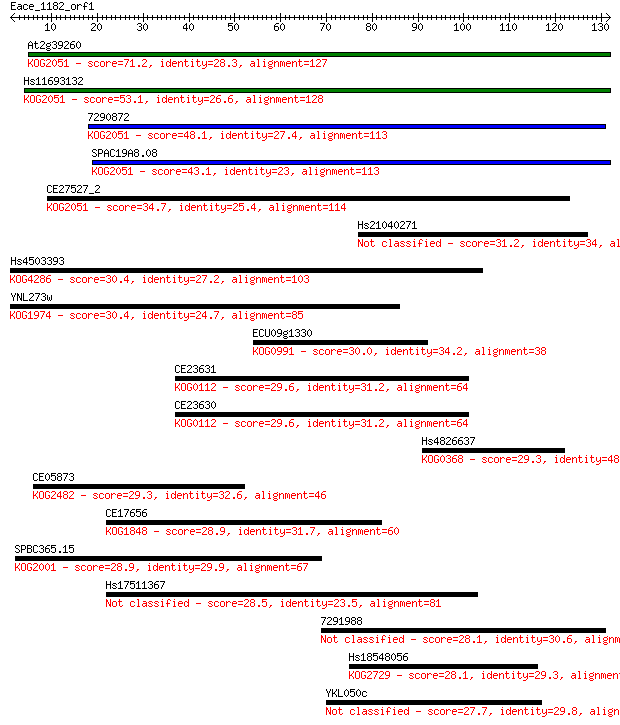
At2g39260 71.2 4e-13
Hs11693132 53.1 1e-07
7290872 48.1 4e-06
SPAC19A8.08 43.1 1e-04
CE27527_2 34.7 0.054
Hs21040271 31.2 0.48
Hs4503393 30.4 0.81
YNL273w 30.4 0.93
ECU09g1330 30.0 1.2
CE23631 29.6 1.5
CE23630 29.6 1.6
Hs4826637 29.3 2.0
CE05873 29.3 2.2
CE17656 28.9 2.5
SPBC365.15 28.9 2.5
Hs17511367 28.5 3.2
7291988 28.1 4.6
Hs18548056 28.1 4.7
YKL050c 27.7 5.7
> At2g39260
Length=1029
Score = 71.2 bits (173), Expect = 4e-13, Method: Composition-based stats.
Identities = 36/127 (28%), Positives = 73/127 (57%), Gaps = 2/127 (1%)
Query 5 LLNRISQATTAQQIDQIAADFFLERLNAKQNRSELIMHILFVAETNVQCMPCCARLLAIL 64
LL R+ + IDQ+ ++ LN+K NR +L+ + V T+++ + +R++A L
Sbjct 313 LLQRLPGCVSRDLIDQLTVEYCY--LNSKTNRKKLVKALFNVPRTSLELLAYYSRMVATL 370
Query 65 RKLLKEETNQALELLQSKWRQLADDCDPTKVEERVRVLRFVCELIKLKLLPPGLLLNCCS 124
+K+ + +++L+ ++ L D +E ++R +RF+ EL K K++P GL+ +C
Sbjct 371 ASCMKDIPSMLVQMLEDEFNSLVHKKDQMNIETKIRNIRFIGELCKFKIVPAGLVFSCLK 430
Query 125 SWLEDFT 131
+ L++FT
Sbjct 431 ACLDEFT 437
> Hs11693132
Length=1272
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 34/128 (26%), Positives = 58/128 (45%), Gaps = 1/128 (0%)
Query 4 ALLNRISQATTAQQIDQIAADFFLERLNAKQNRSELIMHILFVAETNVQCMPCCARLLAI 63
A L ++ ID+ A DF + +N K NR +L+ + V + +P ARL+A
Sbjct 570 AFLQQLPNCVNRDLIDKAAMDFCMN-MNTKANRKKLVRALFIVPRQRLDLLPFYARLVAT 628
Query 64 LRKLLKEETNQALELLQSKWRQLADDCDPTKVEERVRVLRFVCELIKLKLLPPGLLLNCC 123
L + + +L+ +R D +E + + +RF+ EL K K+ L+C
Sbjct 629 LHPCMSDVAEDLCSMLRGDFRFHVRKKDQINIETKNKTVRFIGELTKFKMFTKNDTLHCL 688
Query 124 SSWLEDFT 131
L DF+
Sbjct 689 KMLLSDFS 696
> 7290872
Length=1241
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 31/113 (27%), Positives = 52/113 (46%), Gaps = 1/113 (0%)
Query 18 IDQIAADFFLERLNAKQNRSELIMHILFVAETNVQCMPCCARLLAILRKLLKEETNQALE 77
ID A +F L N K R +L I V T + +P +R +AI+ + E
Sbjct 527 IDSAAIEFLL-NFNTKHQRKKLTRTIFSVQRTRLDILPYLSRFVAIVHMCNTDVATDLAE 585
Query 78 LLQSKWRQLADDCDPTKVEERVRVLRFVCELIKLKLLPPGLLLNCCSSWLEDF 130
LL+ +++ + +E +++++RF+ EL+K L L C L DF
Sbjct 586 LLRKEFKWHIHKKNQLNIETKLKIVRFIGELVKFGLFKKFDALGCLKMLLRDF 638
> SPAC19A8.08
Length=1049
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/113 (23%), Positives = 59/113 (52%), Gaps = 2/113 (1%)
Query 19 DQIAADFFLERLNAKQNRSELIMHILFVAETNVQCMPCCARLLAILRKLLKEETNQALEL 78
+++A +F+ LN K +R+ LI + + T+ +P RL IL +L E + ++
Sbjct 462 NEMALEFY--DLNTKASRNRLIKALCTIPRTSSFLVPYYVRLARILSQLSSEFSTSLVDH 519
Query 79 LQSKWRQLADDCDPTKVEERVRVLRFVCELIKLKLLPPGLLLNCCSSWLEDFT 131
+ ++++ + + R+ ++R++ EL K +L+P ++ C + +FT
Sbjct 520 ARHSFKRMIHRKAKHEYDTRLLIVRYISELTKFQLMPFHMVFECYKLCINEFT 572
> CE27527_2
Length=1098
Score = 34.7 bits (78), Expect = 0.054, Method: Compositional matrix adjust.
Identities = 29/119 (24%), Positives = 56/119 (47%), Gaps = 10/119 (8%)
Query 9 ISQATTAQQIDQIAADFFLERLNAKQNRSELIMHILFVAETNVQCMPCCARLLAILRKLL 68
+S+ +T Q AA +F+ LN K R L+ ++ + + +P ARL+A L ++
Sbjct 402 VSKYSTDQ-----AAIYFVSNLNNKGCRKRLVKLMIDPPPSRIDVVPFYARLVATLENVM 456
Query 69 KEETNQALELLQSKWRQLADDCDPT-----KVEERVRVLRFVCELIKLKLLPPGLLLNC 122
+ T + + L K+R + KVE ++ + + EL+K ++ L+C
Sbjct 457 PDLTTEIVTQLLEKFRGFLQQKPSSATAIIKVESKMVCVMMIAELMKFGVISRAEGLSC 515
> Hs21040271
Length=980
Score = 31.2 bits (69), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 27/50 (54%), Gaps = 1/50 (2%)
Query 77 ELLQSKWRQLADDCDPTKVEERVRVLRFVCELIKLKLLPPGLLLNCCSSW 126
EL+ W +L DD P+ + + R+L V L +LK+ P L+ + C W
Sbjct 225 ELISKDWPELQDDI-PSILAQAQRILFVVDGLDELKVPPGALIQDICGDW 273
> Hs4503393
Length=954
Score = 30.4 bits (67), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 28/103 (27%), Positives = 45/103 (43%), Gaps = 5/103 (4%)
Query 1 AMAALLNRISQATTAQQIDQIAADFFLERLNAKQNRSELIMHILFVAETNVQCMPCCARL 60
AM L +SQA + + D F++ L +L + V+ + A
Sbjct 236 AMEELSTTLSQAEGVRATWEPIGDLFIDSLPEHIQAIKLFKEEFSPMKDGVKLVNDLAHQ 295
Query 61 LAILRKLLKEETNQALELLQSKWRQLADDCDPTKVEERVRVLR 103
LAI L E +QALE + +W+QL V+ER++ L+
Sbjct 296 LAISDVHLSMENSQALEQINVRWKQLQ-----ASVDERLKQLQ 333
> YNL273w
Length=1238
Score = 30.4 bits (67), Expect = 0.93, Method: Composition-based stats.
Identities = 21/86 (24%), Positives = 47/86 (54%), Gaps = 2/86 (2%)
Query 1 AMAALLNRISQATTAQQIDQIAADFFLERLNAKQNRSELIMHILFV-AETNVQCMPCCAR 59
A LLN +S+ AQ+ D +F + RL + + R +L+ ++ + ++ ++Q M C
Sbjct 535 AFNELLNLVSRTKAAQEEDSTDIEFIVSRLFSDE-RIQLLSNLPKIGSKYSLQFMKSCIE 593
Query 60 LLAILRKLLKEETNQALELLQSKWRQ 85
L + K+L++ ++ +++ K R+
Sbjct 594 LTHSVLKVLEQYSDDKTLVIEGKSRR 619
> ECU09g1330
Length=309
Score = 30.0 bits (66), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 54 MPCCARLLAILRKLLKEETNQALELLQSKWRQLADDCD 91
+P R+ +L++LLK E +ALE+ W + D D
Sbjct 219 LPSPKRIEKVLQRLLKREVEEALEMFDEIWEEKFDPLD 256
> CE23631
Length=2722
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 36/65 (55%), Gaps = 3/65 (4%)
Query 37 SELIMHILFVAETNVQCM-PCCARLLAILRKLLKEETNQALELLQSKWRQLADDCDPTKV 95
S + H+L A+ N Q M P +L A +++ N+A +++Q+K +Q + D K
Sbjct 1945 SGIFQHLLLHAQGNGQNMTPEMLQLKAAF--FAQQQENEANQMMQAKMKQQTINKDRIKE 2002
Query 96 EERVR 100
+ERV+
Sbjct 2003 QERVK 2007
> CE23630
Length=2526
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 36/65 (55%), Gaps = 3/65 (4%)
Query 37 SELIMHILFVAETNVQCM-PCCARLLAILRKLLKEETNQALELLQSKWRQLADDCDPTKV 95
S + H+L A+ N Q M P +L A +++ N+A +++Q+K +Q + D K
Sbjct 1749 SGIFQHLLLHAQGNGQNMTPEMLQLKAAF--FAQQQENEANQMMQAKMKQQTINKDRIKE 1806
Query 96 EERVR 100
+ERV+
Sbjct 1807 QERVK 1811
> Hs4826637
Length=2346
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 22/33 (66%), Gaps = 2/33 (6%)
Query 91 DPTKVEERVR--VLRFVCELIKLKLLPPGLLLN 121
DP+K+EE VR V+R+ L KL++L L +N
Sbjct 1484 DPSKIEESVRSMVMRYGSRLWKLRVLQAELKIN 1516
> CE05873
Length=614
Score = 29.3 bits (64), Expect = 2.2, Method: Composition-based stats.
Identities = 15/50 (30%), Positives = 27/50 (54%), Gaps = 4/50 (8%)
Query 6 LNRISQATTAQQIDQIAADFFLE----RLNAKQNRSELIMHILFVAETNV 51
+ R+ +A QQ ++ F L+ R NA+ NRS++I H+ + N+
Sbjct 284 MRRLEEALQCQQREREDTSFQLQCIFCRYNARGNRSKIIHHLYMIHHLNL 333
> CE17656
Length=1646
Score = 28.9 bits (63), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 31/61 (50%), Gaps = 1/61 (1%)
Query 22 AADFFLERLNAKQNRSELIMHILFVAETNVQCMPCCARLLAILRKLLKE-ETNQALELLQ 80
A D + RL +EL H++ + + V MPCC + +L+K ++ Q L LLQ
Sbjct 1579 AVDSLIARLARDPKMTELYSHLVSLFPSVVDVMPCCHADPQLEHQLIKTIKSYQTLFLLQ 1638
Query 81 S 81
+
Sbjct 1639 N 1639
> SPBC365.15
Length=784
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 20/67 (29%), Positives = 33/67 (49%), Gaps = 0/67 (0%)
Query 2 MAALLNRISQATTAQQIDQIAADFFLERLNAKQNRSELIMHILFVAETNVQCMPCCARLL 61
M + + +I TT++ I+ F E NA+ + + HI F+ +CM +RLL
Sbjct 641 MLSFVQKIIYYTTSEVIETHWGKFMGELENARTVDNLMQEHIDFLDTCLKECMLTNSRLL 700
Query 62 AILRKLL 68
+ KLL
Sbjct 701 KVQSKLL 707
> Hs17511367
Length=206
Score = 28.5 bits (62), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 19/84 (22%), Positives = 39/84 (46%), Gaps = 4/84 (4%)
Query 22 AADFFLERLNAKQNRSELIMHILFVAETNVQCMPCCARLLA---ILRKLLKEETNQALEL 78
D L ++ ++ E + +I F + TN++ + +A I R + ++ Q L
Sbjct 23 TTDNMLTDVHIQRQEEEYVRYISFPS-TNIKANKSIQKEIAGNIISRGMQRKVYKQTAGL 81
Query 79 LQSKWRQLADDCDPTKVEERVRVL 102
LQ+ W Q + C T++ R + +
Sbjct 82 LQAVWNQASKQCQNTELSRRTKHI 105
> 7291988
Length=288
Score = 28.1 bits (61), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 33/67 (49%), Gaps = 5/67 (7%)
Query 69 KEETNQALELLQSKWRQLADDCDPTKVEERVRVLRFVCELIKLKLLPP---GLLLNCCSS 125
KE N+A ELL+ K R++ + T+V R+ + R ++++L GL + C
Sbjct 41 KELRNRAYELLERKLREIQPNATRTEVGRRINIFRTNYRREQMRILKQKELGLHSDLCKP 100
Query 126 --WLEDF 130
W D+
Sbjct 101 TLWFYDY 107
> Hs18548056
Length=160
Score = 28.1 bits (61), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 12/41 (29%), Positives = 24/41 (58%), Gaps = 0/41 (0%)
Query 75 ALELLQSKWRQLADDCDPTKVEERVRVLRFVCELIKLKLLP 115
A + L++ ++ D C+P ER+R + +C L++ +LP
Sbjct 30 AFDELRTDFKSPIDQCNPVHARERLRNIERICFLLRKLVLP 70
> YKL050c
Length=922
Score = 27.7 bits (60), Expect = 5.7, Method: Composition-based stats.
Identities = 14/47 (29%), Positives = 21/47 (44%), Gaps = 0/47 (0%)
Query 70 EETNQALELLQSKWRQLADDCDPTKVEERVRVLRFVCELIKLKLLPP 116
E + A L + + DDC T +E++ R F K+ L PP
Sbjct 106 ESVDLAARLASKRTKVSPDDCVETAIEQKARGEAFKVTFSKIPLTPP 152
Lambda K H
0.326 0.136 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1283105810
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40