bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
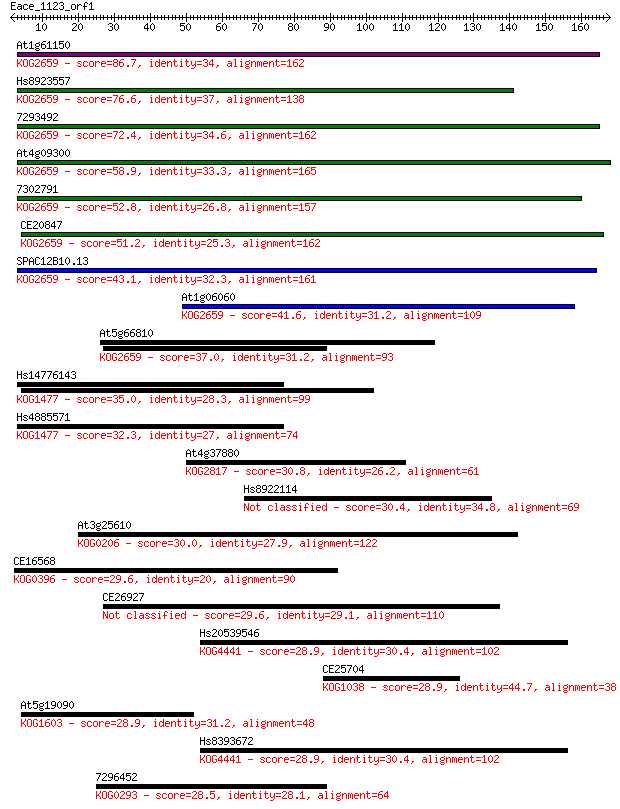
Query= Eace_1123_orf1
Length=167
Score E
Sequences producing significant alignments: (Bits) Value
At1g61150 86.7 2e-17
Hs8923557 76.6 2e-14
7293492 72.4 4e-13
At4g09300 58.9 4e-09
7302791 52.8 3e-07
CE20847 51.2 8e-07
SPAC12B10.13 43.1 2e-04
At1g06060 41.6 7e-04
At5g66810 37.0 0.018
Hs14776143 35.0 0.066
Hs4885571 32.3 0.40
At4g37880 30.8 1.5
Hs8922114 30.4 1.9
At3g25610 30.0 2.3
CE16568 29.6 2.8
CE26927 29.6 3.1
Hs20539546 28.9 4.5
CE25704 28.9 4.6
At5g19090 28.9 4.8
Hs8393672 28.9 5.6
7296452 28.5 7.0
> At1g61150
Length=243
Score = 86.7 bits (213), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 55/163 (33%), Positives = 96/163 (58%), Gaps = 3/163 (1%)
Query 3 AEEFIKEARIDLQMPLHCIDCRAKIREAILSGQTDEAIRQIGFIDPNLLNADAEVTFLLH 62
AE+F +E+ ++ L I R +++A+ +G ++AI ++ ++P +L+ + E+ F L
Sbjct 61 AEKFQRESGTKPEIDLATITDRMAVKKAVQNGNVEDAIEKVNDLNPEILDTNPELFFHLQ 120
Query 63 KQQLLRLIEAGDPEEAIDFAQQRLAPCVKECPHLLPQLEEVMALLAFSDLS-CPEAQRLI 121
+Q+L+ LI G EEA++FAQ+ LAP +E L +LE+ +ALL F D S CP + L
Sbjct 121 QQRLIELIRQGKTEEALEFAQEELAPRGEENQAFLEELEKTVALLVFDDASTCPVKELL- 179
Query 122 GGMEQREETARRIDEAILDLYRIEQESALELLTKKVLFSQGCL 164
+ R +TA ++ AIL E++ L L K ++++Q L
Sbjct 180 -DLSHRLKTASEVNAAILTSQSHEKDPKLPSLLKMLIWAQTQL 221
> Hs8923557
Length=228
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 51/138 (36%), Positives = 77/138 (55%), Gaps = 1/138 (0%)
Query 3 AEEFIKEARIDLQMPLHCIDCRAKIREAILSGQTDEAIRQIGFIDPNLLNADAEVTFLLH 62
AE+F E+ I+ + L +D R KIRE IL GQ EAI I + P LL+ + + F L
Sbjct 46 AEKFRMESGIEPSVDLETLDERIKIREMILKGQIQEAIALINSLHPELLDTNRYLYFHLQ 105
Query 63 KQQLLRLIEAGDPEEAIDFAQQRLAPCVKECPHLLPQLEEVMALLAFSDLSCPEAQRLIG 122
+Q L+ LI + E A++FAQ +LA +E L ++E +ALLAF L+
Sbjct 106 QQHLIELIRQRETEAALEFAQTQLAEQGEESRECLTEMERTLALLAFDSPEESPFGDLLH 165
Query 123 GMEQREETARRIDEAILD 140
M QR++ +++A+LD
Sbjct 166 TM-QRQKVWSEVNQAVLD 182
> 7293492
Length=225
Score = 72.4 bits (176), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 56/165 (33%), Positives = 91/165 (55%), Gaps = 9/165 (5%)
Query 3 AEEFIKEARIDLQMPLHCIDCRAKIREAILSGQTDEAIRQIGFIDPNLLNADAEVTFLLH 62
AE+F EA ++ + L +D R IREA+ +G+ +EA + + + P LL +D + F L
Sbjct 46 AEKFQHEADLEPSVELSSLDGRILIREAVQAGRIEEATQLVNQLHPELLGSDRYLFFHLQ 105
Query 63 KQQLLRLIEAGDPEEAIDFAQQRLAPCVKECPHLLPQLEEVMALLAFSDLSCPEAQRLIG 122
+ QL+ LI AG EEA+ FAQ +L+ +E + +LE +ALLAF P+
Sbjct 106 QLQLIELIRAGKVEEALSFAQSKLSESGEEA---MFELERTLALLAFEK---PQESPFAD 159
Query 123 GMEQ--REETARRIDEAILDLYRIEQES-ALELLTKKVLFSQGCL 164
+EQ R++ A ++ AIL + E + + L K +L++Q L
Sbjct 160 LLEQSYRQKIASELNSAILRCEQSEDSTPKMMFLLKLILWAQSKL 204
> At4g09300
Length=226
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 55/170 (32%), Positives = 90/170 (52%), Gaps = 12/170 (7%)
Query 3 AEEFIKEARIDLQMP---LHCIDCRAKIREAILSGQTDEAIRQIGFIDPNLLNADAEVTF 59
AE+F KE I +Q L ++ R ++ +AI S ++AI ++ ++P ++ + +F
Sbjct 49 AEKF-KEESITMQTAEEDLASMNERLEVIKAIESRNLEDAIEKLNALNPEII----KTSF 103
Query 60 LLHKQQLLRLIEAGDPEEAIDFAQQRLAPCVKECPHLLPQLEEVMALLAFSDL-SCPEAQ 118
LH+Q L+ LI EEA+ FAQ++LAP +E L +LE+ + +L L +CP
Sbjct 104 HLHQQMLIELIREKKTEEAVAFAQEKLAPLAEENEALQRELEKTVCILVTEGLPNCP--S 161
Query 119 RLIGGMEQREETARRIDEAILDLYRIEQESALE-LLTKKVLFSQGCLQNT 167
R + Q TA ++EAI E+ L LLT+KV ++ Q T
Sbjct 162 RELFHNSQWIRTASHVNEAIHTSQTGEKGIWLYFLLTQKVKNNELIYQKT 211
> 7302791
Length=223
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 42/157 (26%), Positives = 73/157 (46%), Gaps = 1/157 (0%)
Query 3 AEEFIKEARIDLQMPLHCIDCRAKIREAILSGQTDEAIRQIGFIDPNLLNADAEVTFLLH 62
A+ F+ EA + + I R +I++A+ GQ A+ I P L D V F +
Sbjct 27 AKRFMTEASVKPGPHMDTIGDRLRIQDAVRVGQVKYAMDLATRIYPRLFETDNYVFFHMQ 86
Query 63 KQQLLRLIEAGDPEEAIDFAQQRLAPCVKECPHLLPQLEEVMALLAFSDLSCPEAQRLIG 122
+ +L+ +I E+A+ FAQ + A K P ++E M LLAF L+
Sbjct 87 QLRLIEMIRDQKMEKALKFAQSKAAGFSKVDPSHYHEVERTMGLLAFDRPEYSPYGELM- 145
Query 123 GMEQREETARRIDEAILDLYRIEQESALELLTKKVLF 159
R++ A ++ A+L + E +S E + +++F
Sbjct 146 YYSYRQKVAGEMNAAMLRCHEDESKSKEEPMEPRMMF 182
> CE20847
Length=234
Score = 51.2 bits (121), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 41/164 (25%), Positives = 83/164 (50%), Gaps = 5/164 (3%)
Query 4 EEFIKEARIDL-QMPLHCIDCRAKIREAILSGQTDEAIRQIGFIDPNLLNADAEVTFLLH 62
E F KE I + + + ++ R ++R I G+ + AI ++ + P +L D E+ F++
Sbjct 28 ETFSKEVNIVVPKKDIDNMNARNEVRRLICVGEMESAIEKMTTLCPTILEDD-EINFIVR 86
Query 63 KQQLLRLIEAGDPEEAIDFAQQRLAPCVKE-CPHLLPQLEEVMALLAFSDLSCPEAQRLI 121
KQ L+ ++ +E +++ + L + C + +E + +L F +L +
Sbjct 87 KQHLIEMVRQKLTKEPVEYFRAHLMKNGQRPCDEKMDIIERIFTMLVF-NLEDDVEFNVY 145
Query 122 GGMEQREETARRIDEAILDLYRIEQESALELLTKKVLFSQGCLQ 165
+RE+TA+ ++ A+L + + S L+LL K + F+ GC Q
Sbjct 146 FQQSEREQTAKEVNSALLAMNGKLKSSRLDLLAKTIAFT-GCSQ 188
> SPAC12B10.13
Length=240
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 52/171 (30%), Positives = 83/171 (48%), Gaps = 14/171 (8%)
Query 3 AEEFIKEARI-DLQMPLHCIDCRAKIREAILSGQTDEAIRQIGFIDPNLLNADAEVTFLL 61
A+ F +EA+I D +P + + R +I E I SG + AI ++ ++P +L+ ++E+ F L
Sbjct 53 AKTFAEEAQITDYYIPPY-VKERLEICELIKSGSINSAICKLNELEPEILDTNSELLFEL 111
Query 62 HKQQLLR----LIEAGDP-----EEAIDFAQQRLAPCVKECPHLLPQLEEVMALLAFSDL 112
+ +LL ++E D E ++FA + LAP L LE M+LL F
Sbjct 112 LRLRLLELIREVVEEKDTSDLAVERCLNFAHENLAPLAPSNQKFLNSLELTMSLLCFPPS 171
Query 113 SCPEAQRLIGGMEQREETARRIDEAILDLYRIEQESALELLTKKVLFSQGC 163
S A + + QRE A + +IL + ES L L V F + C
Sbjct 172 SYSPALKNVLNYSQRERVANLANVSILKSQGLSNESRLLSL---VNFERWC 219
> At1g06060
Length=213
Score = 41.6 bits (96), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 34/111 (30%), Positives = 57/111 (51%), Gaps = 6/111 (5%)
Query 49 NLLNADAEVTFLLHKQQLLRLIEAGDPEEAIDFAQQRLAPCVKECPHLLPQLEEVMALLA 108
+LL + ++ F L + LI AG+ EA+ F + RLAP K + +LE+VMALLA
Sbjct 80 DLLEKNKDLQFDLLCLHFVELICAGNCTEALKFGKTRLAPFGK-VKKYVEKLEDVMALLA 138
Query 109 FSDLSCPEAQRLIG--GMEQREETARRIDEAILDLYRIEQESALELLTKKV 157
+ D PE + E R++ A ++ IL+ + +E + ++V
Sbjct 139 YED---PEKSPMFHLLSSEYRQQVADNLNRTILEHTNHPSYTPMERIIQQV 186
> At5g66810
Length=752
Score = 37.0 bits (84), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 29/96 (30%), Positives = 47/96 (48%), Gaps = 4/96 (4%)
Query 26 KIREAILSGQTDEAIRQIGFIDPNLLNADAEVTFLLHKQQLLRLIEAGDPEEAIDFAQQR 85
+IR +I SG + AI + P +L+ D + F L KQ+ + L+ G E AID +
Sbjct 119 RIRSSIESGDIETAIDILRSHAPFVLD-DHRILFRLQKQKFIELLRKGTHEAAIDCLRTC 177
Query 86 LAPCVKEC-PHLLPQLEEVMALLAF--SDLSCPEAQ 118
+APC + P + + V+ L + D + P A
Sbjct 178 VAPCALDAYPEAYEEFKHVLLALIYDKDDQTSPVAN 213
Score = 37.0 bits (84), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 18/62 (29%), Positives = 31/62 (50%), Gaps = 0/62 (0%)
Query 27 IREAILSGQTDEAIRQIGFIDPNLLNADAEVTFLLHKQQLLRLIEAGDPEEAIDFAQQRL 86
++E + G EA +I +DP+ + + F L + + L+L+ AGD A+ A L
Sbjct 504 LKELVSRGMAAEAFSEISTMDPDFFTQNPGLLFHLKQVEFLKLVSAGDHNGALKVACFHL 563
Query 87 AP 88
P
Sbjct 564 GP 565
> Hs14776143
Length=503
Score = 35.0 bits (79), Expect = 0.066, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 33/74 (44%), Gaps = 0/74 (0%)
Query 3 AEEFIKEARIDLQMPLHCIDCRAKIREAILSGQTDEAIRQIGFIDPNLLNADAEVTFLLH 62
A F + +Q I R KI++ +L G+ EAI P LL + + F+L
Sbjct 157 ATAFARMTETPIQEEQASIKNRQKIQKLVLEGRVGEAIETTQRFYPGLLEHNPNLLFMLK 216
Query 63 KQQLLRLIEAGDPE 76
+Q + ++ D E
Sbjct 217 CRQFVEMVNGTDSE 230
Score = 28.1 bits (61), Expect = 9.1, Method: Composition-based stats.
Identities = 24/107 (22%), Positives = 48/107 (44%), Gaps = 9/107 (8%)
Query 4 EEFIKEARIDLQMPLHCIDCRAKIRE--AILSGQTDEAIRQIGFIDPNLLNA-------D 54
E++++E R +Q +HC A++ E A+L + G+ A
Sbjct 110 EDYMREWRAKVQGTVHCFPISARLGEWQAVLQNMVSSYLVHHGYCATATAFARMTETPIQ 169
Query 55 AEVTFLLHKQQLLRLIEAGDPEEAIDFAQQRLAPCVKECPHLLPQLE 101
E + ++Q++ +L+ G EAI+ Q+ ++ P+LL L+
Sbjct 170 EEQASIKNRQKIQKLVLEGRVGEAIETTQRFYPGLLEHNPNLLFMLK 216
> Hs4885571
Length=500
Score = 32.3 bits (72), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 20/74 (27%), Positives = 36/74 (48%), Gaps = 0/74 (0%)
Query 3 AEEFIKEARIDLQMPLHCIDCRAKIREAILSGQTDEAIRQIGFIDPNLLNADAEVTFLLH 62
AE F + + L I R +I++ +L+G+ EAI + P+LL + + F L
Sbjct 157 AEAFARSTDQTVLEELASIKNRQRIQKLVLAGRMGEAIETTQQLYPSLLERNPNLLFTLK 216
Query 63 KQQLLRLIEAGDPE 76
+Q + ++ D E
Sbjct 217 VRQFIEMVNGTDSE 230
> At4g37880
Length=388
Score = 30.8 bits (68), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 16/61 (26%), Positives = 33/61 (54%), Gaps = 3/61 (4%)
Query 50 LLNADAEVTFLLHKQQLLRLIEAGDPEEAIDFAQQRLAPCVKECPHLLPQLEEVMALLAF 109
L A +++ LH L + + +EAID+A++ +A C LP+++++M L +
Sbjct 180 LKEARSDLEMKLHSLHFLEIARGKNSKEAIDYARKHIATFADSC---LPEIQKLMCSLLW 236
Query 110 S 110
+
Sbjct 237 N 237
> Hs8922114
Length=1069
Score = 30.4 bits (67), Expect = 1.9, Method: Composition-based stats.
Identities = 24/77 (31%), Positives = 36/77 (46%), Gaps = 9/77 (11%)
Query 66 LLRLIEAGDPEEAIDFAQQRLAP-----CVKECPHLL---PQLEEVMALLAFSDLSCPEA 117
L L E+ E + F + RL P +KE PQ E++ LAF + +CP
Sbjct 613 LFSLAESRKKTERLAFRKNRLPPEFSPVIIKEWAAYKGKSPQTPELVEALAFREWTCPNL 672
Query 118 QRLIGGMEQREETARRI 134
+RL G + E+ RR+
Sbjct 673 KRLWLG-KAVEDKNRRM 688
> At3g25610
Length=1202
Score = 30.0 bits (66), Expect = 2.3, Method: Composition-based stats.
Identities = 34/124 (27%), Positives = 51/124 (41%), Gaps = 12/124 (9%)
Query 20 CID--CRAKIREAILSGQTDEAIRQIGFIDPNLLNADAEVTFLLHKQQLLRLIEAGDPEE 77
CID +A I+ +L+G E IGF L ++ L Q+ L ++G +E
Sbjct 715 CIDKLAQAGIKIWVLTGDKMETAINIGFASSLLRQEMKQIIINLETPQIKSLEKSGGKDE 774
Query 78 AIDFAQQRLAPCVKECPHLLPQLEEVMALLAFSDLSCPEAQRLIGGMEQREETARRIDEA 137
I+ A + ++ QL+E ALLA S S +I G I +
Sbjct 775 -IELASRE---------SVVMQLQEGKALLAASGASSEAFALIIDGKSLTYALEDEIKKM 824
Query 138 ILDL 141
LDL
Sbjct 825 FLDL 828
> CE16568
Length=428
Score = 29.6 bits (65), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 18/90 (20%), Positives = 43/90 (47%), Gaps = 0/90 (0%)
Query 2 PAEEFIKEARIDLQMPLHCIDCRAKIREAILSGQTDEAIRQIGFIDPNLLNADAEVTFLL 61
PA+ +KE ++ + + + +++A+L G + L ++ + +
Sbjct 169 PAKVLVKEMELEDLVDVDVFENMYAVQQALLDGNIQPCLAWCDRHHRKLRKLESRIELVA 228
Query 62 HKQQLLRLIEAGDPEEAIDFAQQRLAPCVK 91
+Q+ + LIE G+ EA+ + ++ +AP K
Sbjct 229 RQQEAVTLIELGNIPEAVAYVKKYIAPIAK 258
> CE26927
Length=290
Score = 29.6 bits (65), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 32/118 (27%), Positives = 53/118 (44%), Gaps = 10/118 (8%)
Query 27 IREAILSGQTDEAIRQIGFIDP-NLLNADAEVTFLLHKQQLLRLI----EAGDPEEAIDF 81
+R+ IL GQ D A+ + + P N + A + + K + L+ E G P DF
Sbjct 69 LRQLILDGQWDNALDFVEPLKPVNEFDFGA-FRYNITKYKFFELLCVKLEPG-PLHDNDF 126
Query 82 AQQRLAPCVKECPHLLPQLEE---VMALLAFSDLSCPEAQRLIGGMEQREETARRIDE 136
A + L C+K+ H+ P E+ + ALL LS E + R E ++++
Sbjct 127 AVEELVECLKDLEHICPTPEDYRHLCALLTLPKLSDHEDFKNWNPSSARVECFNKVNQ 184
> Hs20539546
Length=587
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 31/120 (25%), Positives = 47/120 (39%), Gaps = 20/120 (16%)
Query 54 DAEVTFLLHKQQLLRLIEAGDPEEAIDFAQQRLAPC----------VKECPHLLPQ---- 99
+ V LL LL+L++ + DF Q +L P V C LL Q
Sbjct 117 EENVQVLLPAASLLQLMDV--RQNCCDFLQSQLHPTNCLGIRAFADVHTCTDLLQQANAY 174
Query 100 ----LEEVMALLAFSDLSCPEAQRLIGGMEQREETARRIDEAILDLYRIEQESALELLTK 155
EVM F LS + LI + + ++ EA++ E+E+ LE + K
Sbjct 175 AEQHFPEVMLGEEFLSLSLDQVCSLISSDKLTVSSEEKVFEAVISWINYEKETRLEHMAK 234
> CE25704
Length=1154
Score = 28.9 bits (63), Expect = 4.6, Method: Composition-based stats.
Identities = 17/39 (43%), Positives = 23/39 (58%), Gaps = 2/39 (5%)
Query 88 PCVKECPHLLPQLEEVMALLAFSDLS-CPEAQRLIGGME 125
P V C LLP+ E+++ L FS LS C + Q LIG +
Sbjct 354 PIVSTCLELLPR-EKLVETLHFSALSACSQGQNLIGASQ 391
> At5g19090
Length=587
Score = 28.9 bits (63), Expect = 4.8, Method: Composition-based stats.
Identities = 15/57 (26%), Positives = 31/57 (54%), Gaps = 9/57 (15%)
Query 4 EEFIKEARIDLQMPLHCIDCRAKIR------EAILSGQTDEAIRQI---GFIDPNLL 51
EEF+K L++ +HC C+ K++ E + + + D + ++ G +DP++L
Sbjct 4 EEFMKIQTCVLKVNIHCDGCKQKVKKILQKIEGVFTTKIDAELGKVTVSGNVDPSVL 60
> Hs8393672
Length=587
Score = 28.9 bits (63), Expect = 5.6, Method: Composition-based stats.
Identities = 31/120 (25%), Positives = 47/120 (39%), Gaps = 20/120 (16%)
Query 54 DAEVTFLLHKQQLLRLIEAGDPEEAIDFAQQRLAPC----------VKECPHLLPQ---- 99
+ V LL LL+L++ + DF Q +L P V C LL Q
Sbjct 117 EENVQVLLPAASLLQLMDV--RQNCCDFLQSQLHPTNCLGIRAFADVHTCTDLLQQANAY 174
Query 100 ----LEEVMALLAFSDLSCPEAQRLIGGMEQREETARRIDEAILDLYRIEQESALELLTK 155
EVM F LS + LI + + ++ EA++ E+E+ LE + K
Sbjct 175 AEQHFPEVMLGEEFLSLSLDQVCSLISSDKLTVSSEEKVFEAVISWINYEKENRLEHMAK 234
> 7296452
Length=324
Score = 28.5 bits (62), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 18/68 (26%), Positives = 34/68 (50%), Gaps = 4/68 (5%)
Query 25 AKIREAILSGQTDEAIRQIGFIDPNLLNAD----AEVTFLLHKQQLLRLIEAGDPEEAID 80
K RE +L G +A + ++P + N E+ F+L +Q+ L ++ G+P +A+
Sbjct 134 TKFREHVLMGDWSKADSDLKDLEPLIDNGKLATITEMKFILLEQKYLEHLDDGNPLDALH 193
Query 81 FAQQRLAP 88
+ L P
Sbjct 194 VLRSELTP 201
Lambda K H
0.322 0.138 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2425968562
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40