bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1119_orf1
Length=150
Score E
Sequences producing significant alignments: (Bits) Value
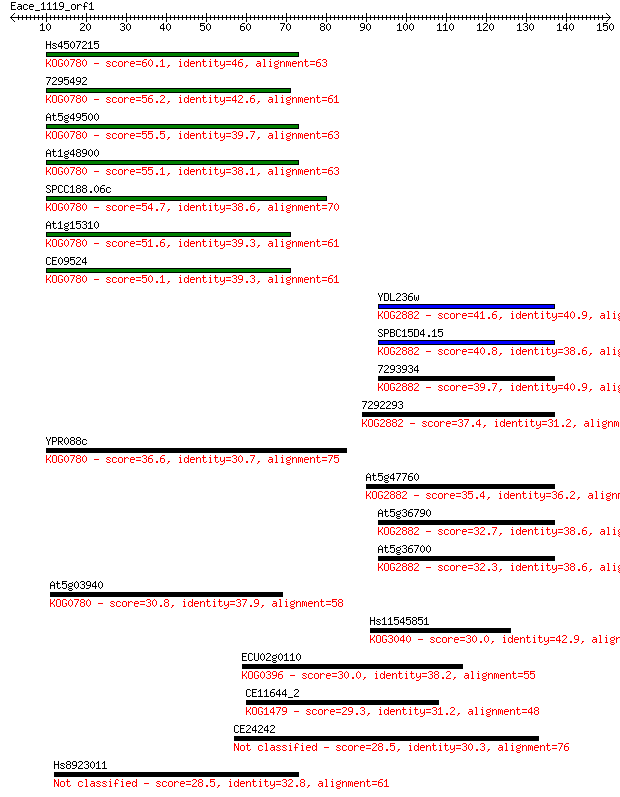
Hs4507215 60.1 2e-09
7295492 56.2 2e-08
At5g49500 55.5 3e-08
At1g48900 55.1 5e-08
SPCC188.06c 54.7 6e-08
At1g15310 51.6 5e-07
CE09524 50.1 1e-06
YDL236w 41.6 6e-04
SPBC15D4.15 40.8 0.001
7293934 39.7 0.002
7292293 37.4 0.012
YPR088c 36.6 0.019
At5g47760 35.4 0.041
At5g36790 32.7 0.27
At5g36700 32.3 0.33
At5g03940 30.8 1.1
Hs11545851 30.0 1.6
ECU02g0110 30.0 1.9
CE11644_2 29.3 2.6
CE24242 28.5 5.1
Hs8923011 28.5 5.5
> Hs4507215
Length=504
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 29/63 (46%), Positives = 47/63 (74%), Gaps = 0/63 (0%)
Query 10 MVLAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQI 69
MVLA+LG++I+ ALR+L +ATI++E V+N +K VC ALL ADV +++V++ + V I
Sbjct 1 MVLADLGRKITSALRSLSNATIINEEVLNAMLKEVCTALLEADVNIKLVKQLRENVKSAI 60
Query 70 NTQ 72
+ +
Sbjct 61 DLE 63
> 7295492
Length=508
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 26/61 (42%), Positives = 43/61 (70%), Gaps = 0/61 (0%)
Query 10 MVLAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQI 69
MVLA+LG++I+ AL +L AT+++E +N +K +C ALL ADV +R+V++ + V I
Sbjct 1 MVLADLGRKITTALHSLSKATVINEEALNSMLKEICAALLEADVNIRLVKQLRENVRAVI 60
Query 70 N 70
+
Sbjct 61 D 61
> At5g49500
Length=497
Score = 55.5 bits (132), Expect = 3e-08, Method: Composition-based stats.
Identities = 25/63 (39%), Positives = 42/63 (66%), Gaps = 0/63 (0%)
Query 10 MVLAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQI 69
MVLAELG +I A++ + + TI+DE +NDC+ + RALL +DV +V+E + + + +
Sbjct 1 MVLAELGGRIMSAIQKMNNVTIIDEKALNDCLNEITRALLQSDVSFPLVKEMQTNIKKIV 60
Query 70 NTQ 72
N +
Sbjct 61 NLE 63
> At1g48900
Length=522
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 24/63 (38%), Positives = 43/63 (68%), Gaps = 0/63 (0%)
Query 10 MVLAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQI 69
MVLAELG +I+ A++ + + TI+DE +N+C+ + RALL +DV +V+E + + + +
Sbjct 1 MVLAELGGRITRAIQQMSNVTIIDEKALNECLNEITRALLQSDVSFPLVKEMQSNIKKIV 60
Query 70 NTQ 72
N +
Sbjct 61 NLE 63
> SPCC188.06c
Length=522
Score = 54.7 bits (130), Expect = 6e-08, Method: Composition-based stats.
Identities = 27/70 (38%), Positives = 44/70 (62%), Gaps = 0/70 (0%)
Query 10 MVLAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQI 69
MV A+LG++++ AL + AT V+E +V+ +K +C ALL DV +R+VQE + + ++I
Sbjct 1 MVFADLGRRLNSALGDFSKATSVNEELVDTLLKNICTALLETDVNVRLVQELRSNIKKKI 60
Query 70 NTQIRQQSSN 79
N Q N
Sbjct 61 NVSTLPQGIN 70
> At1g15310
Length=479
Score = 51.6 bits (122), Expect = 5e-07, Method: Composition-based stats.
Identities = 24/61 (39%), Positives = 42/61 (68%), Gaps = 0/61 (0%)
Query 10 MVLAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQI 69
MVLAELG +I+ A++ + + TI+DE V+ND + + RALL +DV +V++ + + + +
Sbjct 1 MVLAELGGRITRAIQQMNNVTIIDEKVLNDFLNEITRALLQSDVSFGLVEKMQTNIKKIV 60
Query 70 N 70
N
Sbjct 61 N 61
> CE09524
Length=496
Score = 50.1 bits (118), Expect = 1e-06, Method: Composition-based stats.
Identities = 24/61 (39%), Positives = 42/61 (68%), Gaps = 0/61 (0%)
Query 10 MVLAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQI 69
MVLA+LG++I A+ L +T+++E ++ +K VC AL+ +DV +R+V++ K V + I
Sbjct 1 MVLADLGRKIRNAIGKLGQSTVINEGELDLMLKEVCTALIESDVHIRLVKQLKDNVKKAI 60
Query 70 N 70
N
Sbjct 61 N 61
> YDL236w
Length=312
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 27/44 (61%), Gaps = 0/44 (0%)
Query 93 KSVYFLTNNSTLSRKGFVAKLNKLGIEATEQQVLCTSYSTARYI 136
K + F+TNNST SR + K GI+ E+Q+ + Y++A YI
Sbjct 57 KQLIFVTNNSTKSRLAYTKKFASFGIDVKEEQIFTSGYASAVYI 100
> SPBC15D4.15
Length=298
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 32/44 (72%), Gaps = 0/44 (0%)
Query 93 KSVYFLTNNSTLSRKGFVAKLNKLGIEATEQQVLCTSYSTARYI 136
K + F++NNST SR+ ++ K+N+ GI A +++ ++YS+A Y+
Sbjct 51 KQIIFVSNNSTKSRETYMNKINEHGIAAKLEEIYPSAYSSATYV 94
> 7293934
Length=330
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 28/44 (63%), Gaps = 0/44 (0%)
Query 93 KSVYFLTNNSTLSRKGFVAKLNKLGIEATEQQVLCTSYSTARYI 136
KS+YF TNNST +R + K +LG E ++ T+++TA Y+
Sbjct 72 KSIYFCTNNSTKTRSELLKKGVELGFHIKENGIISTAHATAAYL 115
> 7292293
Length=592
Score = 37.4 bits (85), Expect = 0.012, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 29/48 (60%), Gaps = 0/48 (0%)
Query 89 SSSKKSVYFLTNNSTLSRKGFVAKLNKLGIEATEQQVLCTSYSTARYI 136
+++ + ++ ++NNS +SR+ K GIE E VL +S+S A ++
Sbjct 338 NTTGRKIFIISNNSEISRQEMADKAKGFGIEIKEDNVLTSSFSCANFL 385
> YPR088c
Length=541
Score = 36.6 bits (83), Expect = 0.019, Method: Composition-based stats.
Identities = 23/76 (30%), Positives = 45/76 (59%), Gaps = 1/76 (1%)
Query 10 MVLAELGQQISGALRNLQSATIVD-EAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQ 68
MVLA+LG++I+ A+ N S T D V+ +K + ALL +DV + +V + + + Q
Sbjct 1 MVLADLGKRINSAVNNAISNTQDDFTTSVDVMLKGIVTALLESDVNIALVSKLRNNIRSQ 60
Query 69 INTQIRQQSSNSSSSS 84
+ ++ R + S +++ +
Sbjct 61 LLSENRSEKSTTNAQT 76
> At5g47760
Length=301
Score = 35.4 bits (80), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 30/48 (62%), Gaps = 1/48 (2%)
Query 90 SSKKSVYFLTNNSTLSRKGFVAKLNKLGIEA-TEQQVLCTSYSTARYI 136
S K+V F+TNNS SR+ + K LG+ + T+ ++ +S++ A Y+
Sbjct 48 SKGKNVVFVTNNSVKSRRQYAEKFRSLGVTSITQDEIFSSSFAAAMYL 95
> At5g36790
Length=362
Score = 32.7 bits (73), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 28/44 (63%), Gaps = 0/44 (0%)
Query 93 KSVYFLTNNSTLSRKGFVAKLNKLGIEATEQQVLCTSYSTARYI 136
K + F+TNNST SRK + K LG+ E+++ +S++ A Y+
Sbjct 112 KRLVFVTNNSTKSRKQYGKKFETLGLNVNEEEIFASSFAAAAYL 155
> At5g36700
Length=289
Score = 32.3 bits (72), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 28/44 (63%), Gaps = 0/44 (0%)
Query 93 KSVYFLTNNSTLSRKGFVAKLNKLGIEATEQQVLCTSYSTARYI 136
K + F+TNNST SRK + K LG+ E+++ +S++ A Y+
Sbjct 59 KRLVFVTNNSTKSRKQYGKKFETLGLNVNEEEIFASSFAAAAYL 102
> At5g03940
Length=564
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 22/62 (35%), Positives = 33/62 (53%), Gaps = 4/62 (6%)
Query 11 VLAELGQQISGALRNLQSATIVDEAVVNDCV----KTVCRALLLADVQLRVVQEFKRKVT 66
V AE+ Q++G L S +E + D + + + RALL ADV L VV+ F + V+
Sbjct 73 VRAEMFGQLTGGLEAAWSKLKGEEVLTKDNIAEPMRDIRRALLEADVSLPVVRRFVQSVS 132
Query 67 QQ 68
Q
Sbjct 133 DQ 134
> Hs11545851
Length=270
Score = 30.0 bits (66), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 91 SKKSVYFLTNNSTLSRKGFVAKLNKLGIEATEQQV 125
S+ V F TN S SR V +L +LG + +EQ+V
Sbjct 46 SRLKVRFCTNESQKSRAELVGQLQRLGFDISEQEV 80
> ECU02g0110
Length=336
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 21/55 (38%), Positives = 30/55 (54%), Gaps = 1/55 (1%)
Query 59 QEFKRKVTQQINTQIRQQSSNSSSSSSSSSSSSKKSVYFLTNNSTLSRKGFVAKL 113
+EFKR V +QI + + SNSS SS SK + FL+ L R+G ++L
Sbjct 181 KEFKR-VPEQIKSYLPVLVSNSSFRKYPSSGHSKTAEQFLSCALKLFRRGIKSRL 234
> CE11644_2
Length=737
Score = 29.3 bits (64), Expect = 2.6, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 27/48 (56%), Gaps = 0/48 (0%)
Query 60 EFKRKVTQQINTQIRQQSSNSSSSSSSSSSSSKKSVYFLTNNSTLSRK 107
E KR+V + T+ R++ +N SSS ++S K+++ F T+ K
Sbjct 61 EKKRQVLRNWITRKRREFNNEDSSSQTTSKIPKRAILFCVMTDTVKEK 108
> CE24242
Length=527
Score = 28.5 bits (62), Expect = 5.1, Method: Composition-based stats.
Identities = 23/76 (30%), Positives = 43/76 (56%), Gaps = 7/76 (9%)
Query 57 VVQEFKRKVTQQINTQIRQQSSNSSSSSSSSSSSSKKSVYFLTNNSTLSRKGFVAKLNKL 116
++ + KRK+TQ N +++ ++ S +S+ S++SKKS ST S K F +KL
Sbjct 207 LMSKMKRKITQAKNGH--KKAEDTMSMASAWSATSKKSTI-----STKSGKFFGKIGSKL 259
Query 117 GIEATEQQVLCTSYST 132
G++ E+ + ++ T
Sbjct 260 GLKDKEKPLPVGTFLT 275
> Hs8923011
Length=294
Score = 28.5 bits (62), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 35/61 (57%), Gaps = 12/61 (19%)
Query 12 LAELGQQISGALRNLQSATIVDEAVVNDCVKTVCRALLLADVQLRVVQEFKRKVTQQINT 71
LA + +Q++GALR L+ + + VK + + V+L V+QE KR++T Q+ +
Sbjct 185 LAHVREQMAGALRKLRQ--------LEEQVKLIP----VLQVKLSVLQEEKRQLTVQLKS 232
Query 72 Q 72
Q
Sbjct 233 Q 233
Lambda K H
0.312 0.120 0.307
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1852391706
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40