bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
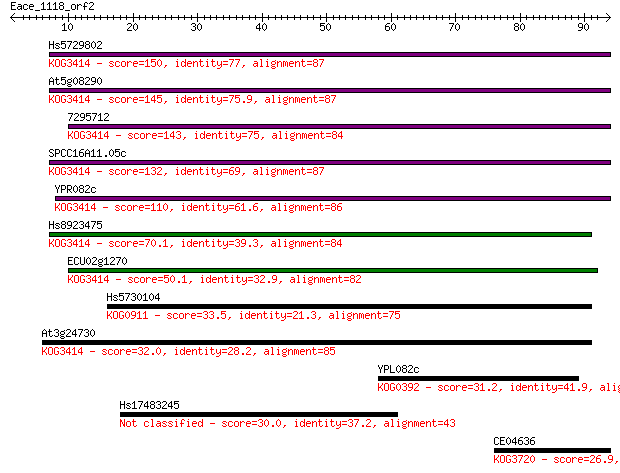
Query= Eace_1118_orf2
Length=93
Score E
Sequences producing significant alignments: (Bits) Value
Hs5729802 150 5e-37
At5g08290 145 1e-35
7295712 143 7e-35
SPCC16A11.05c 132 2e-31
YPR082c 110 8e-25
Hs8923475 70.1 9e-13
ECU02g1270 50.1 9e-07
Hs5730104 33.5 0.099
At3g24730 32.0 0.25
YPL082c 31.2 0.46
Hs17483245 30.0 1.1
CE04636 26.9 9.1
> Hs5729802
Length=142
Score = 150 bits (379), Expect = 5e-37, Method: Compositional matrix adjust.
Identities = 67/87 (77%), Positives = 77/87 (88%), Gaps = 0/87 (0%)
Query 7 MSYMLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYV 66
MSYML HLH+GWQVDQAIL+EE+R+V IRFGHD+DP CMKMDE+L+ +AE VKNF VIY+
Sbjct 1 MSYMLPHLHNGWQVDQAILSEEDRVVVIRFGHDWDPTCMKMDEVLYSIAEKVKNFAVIYL 60
Query 67 VDTTEVPDFTTMYELYDPVTVMFFYRN 93
VD TEVPDF MYELYDP TVMFF+RN
Sbjct 61 VDITEVPDFNKMYELYDPCTVMFFFRN 87
> At5g08290
Length=142
Score = 145 bits (367), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 66/87 (75%), Positives = 74/87 (85%), Gaps = 0/87 (0%)
Query 7 MSYMLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYV 66
MSY+L HLHSGW VDQ+IL EEERLV IRFGHD+D CM+MDE+L VAE +KNF VIY+
Sbjct 1 MSYLLPHLHSGWAVDQSILAEEERLVVIRFGHDWDETCMQMDEVLASVAETIKNFAVIYL 60
Query 67 VDTTEVPDFTTMYELYDPVTVMFFYRN 93
VD TEVPDF TMYELYDP TVMFF+RN
Sbjct 61 VDITEVPDFNTMYELYDPSTVMFFFRN 87
> 7295712
Length=139
Score = 143 bits (361), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 63/84 (75%), Positives = 74/84 (88%), Gaps = 0/84 (0%)
Query 10 MLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYVVDT 69
ML HLH+GWQVDQAIL+EE+R+V IRFGHD+DP CMKMDE+++ +AE VKNF VIY+VD
Sbjct 1 MLPHLHNGWQVDQAILSEEDRVVVIRFGHDWDPACMKMDEVMYSIAEKVKNFAVIYLVDI 60
Query 70 TEVPDFTTMYELYDPVTVMFFYRN 93
TEVPDF MYELYDP TVMFF+RN
Sbjct 61 TEVPDFNKMYELYDPCTVMFFFRN 84
> SPCC16A11.05c
Length=142
Score = 132 bits (332), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 60/87 (68%), Positives = 69/87 (79%), Gaps = 0/87 (0%)
Query 7 MSYMLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYV 66
MSY L HLHSGW VDQAIL+E+ERLV IRFG D D EC+K DE+L+++AE V N VIY+
Sbjct 1 MSYFLPHLHSGWHVDQAILSEQERLVVIRFGRDHDEECIKQDEVLYRIAEKVVNMAVIYL 60
Query 67 VDTTEVPDFTTMYELYDPVTVMFFYRN 93
VD EVPDF MYELYD T+MFFYRN
Sbjct 61 VDIDEVPDFNKMYELYDRTTIMFFYRN 87
> YPR082c
Length=143
Score = 110 bits (274), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 53/86 (61%), Positives = 62/86 (72%), Gaps = 0/86 (0%)
Query 8 SYMLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYVV 67
S +L L +GW VDQAI+TE +RLV IRFG D +CM MDELL +AE V+NF VIY+
Sbjct 3 SVLLPQLRTGWHVDQAIVTETKRLVVIRFGRKNDRQCMIMDELLSSIAERVRNFAVIYLC 62
Query 68 DTTEVPDFTTMYELYDPVTVMFFYRN 93
D EV DF MYEL DP+TVMFFY N
Sbjct 63 DIDEVSDFDEMYELTDPMTVMFFYHN 88
> Hs8923475
Length=149
Score = 70.1 bits (170), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 33/85 (38%), Positives = 54/85 (63%), Gaps = 1/85 (1%)
Query 7 MSYMLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYV 66
MS++L L S +VDQAI + E+++ +RFG D DP C+++D++L K + D+ IY+
Sbjct 1 MSFLLPKLTSKKEVDQAIKSTAEKVLVLRFGRDEDPVCLQLDDILSKTSSDLSKMAAIYL 60
Query 67 VDTTEVPDFTTMYEL-YDPVTVMFF 90
VD + +T +++ Y P TV FF
Sbjct 61 VDVDQTAVYTQYFDISYIPSTVFFF 85
> ECU02g1270
Length=140
Score = 50.1 bits (118), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 42/82 (51%), Gaps = 0/82 (0%)
Query 10 MLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYVVDT 69
M+ L S V+ A+ +LV +RFG DP C+ MD LL ++ + N+ IYV +
Sbjct 1 MINTLESLDAVNGAVGGTSCKLVVVRFGDRGDPLCIHMDGLLERICLALSNYVEIYVCER 60
Query 70 TEVPDFTTMYELYDPVTVMFFY 91
+ V + L PV +M F+
Sbjct 61 SSVRELVDPMGLDSPVNIMCFF 82
> Hs5730104
Length=335
Score = 33.5 bits (75), Expect = 0.099, Method: Composition-based stats.
Identities = 16/75 (21%), Positives = 41/75 (54%), Gaps = 1/75 (1%)
Query 16 SGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYVVDTTEVPDF 75
S Q ++ + + + L+ + F + P+C +M+E++ ++A+++ + ++ VP+
Sbjct 18 SAGQFEELLRLKAKSLLVVHFWAPWAPQCAQMNEVMAELAKELPQVSFVK-LEAEGVPEV 76
Query 76 TTMYELYDPVTVMFF 90
+ YE+ T +FF
Sbjct 77 SEKYEISSVPTFLFF 91
> At3g24730
Length=151
Score = 32.0 bits (71), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 24/89 (26%), Positives = 41/89 (46%), Gaps = 13/89 (14%)
Query 6 KMSYMLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIY 65
+MSY+L+ L + ++D+ I + ++ +RFG D L K DV F +
Sbjct 8 EMSYLLKTLTTKEEIDRVIRDTIDEVLVLRFGRSSDA--------LAKSVRDVSKFAKVA 59
Query 66 VVDTTEVPDFTTMYELYD----PVTVMFF 90
+VD + D + +D P T+ FF
Sbjct 60 LVD-VDSEDVQVYVKYFDITLFPSTIFFF 87
> YPL082c
Length=1867
Score = 31.2 bits (69), Expect = 0.46, Method: Composition-based stats.
Identities = 13/31 (41%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 58 VKNFCVIYVVDTTEVPDFTTMYELYDPVTVM 88
VKN C VDT+EVPDF+ E + + +
Sbjct 979 VKNLCGFLCVDTSEVPDFSVNAEYKEKILTL 1009
> Hs17483245
Length=623
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 23/44 (52%), Gaps = 1/44 (2%)
Query 18 WQVDQAILTEEERLVCIRF-GHDFDPECMKMDELLFKVAEDVKN 60
W QA+L + LVC+R G DPEC+ ++ E +KN
Sbjct 313 WAQVQALLLPDAPLVCVRTDGEGEDPECLGEGKMENPKGESLKN 356
> CE04636
Length=344
Score = 26.9 bits (58), Expect = 9.1, Method: Composition-based stats.
Identities = 10/18 (55%), Positives = 13/18 (72%), Gaps = 0/18 (0%)
Query 76 TTMYELYDPVTVMFFYRN 93
T ++ELYD TV FY+N
Sbjct 278 TVLFELYDDNTVQLFYKN 295
Lambda K H
0.326 0.140 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167934574
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40