bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
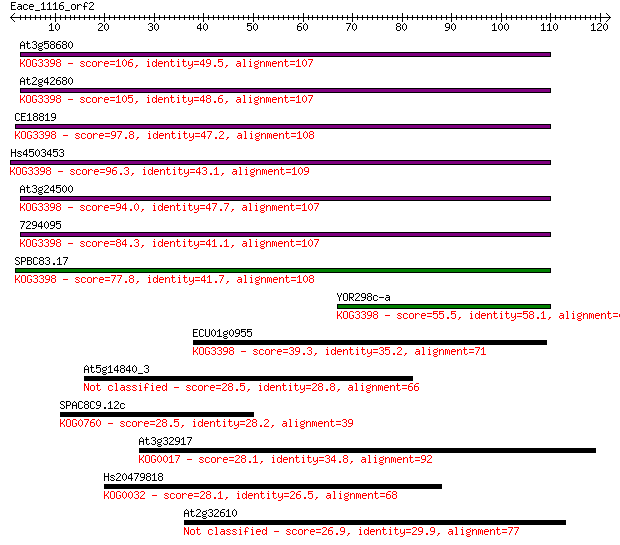
Query= Eace_1116_orf2
Length=121
Score E
Sequences producing significant alignments: (Bits) Value
At3g58680 106 1e-23
At2g42680 105 3e-23
CE18819 97.8 4e-21
Hs4503453 96.3 1e-20
At3g24500 94.0 7e-20
7294095 84.3 5e-17
SPBC83.17 77.8 5e-15
YOR298c-a 55.5 3e-08
ECU01g0955 39.3 0.002
At5g14840_3 28.5 3.3
SPAC8C9.12c 28.5 3.5
At3g32917 28.1 4.1
Hs20479818 28.1 4.4
At2g32610 26.9 8.3
> At3g58680
Length=142
Score = 106 bits (265), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 53/107 (49%), Positives = 72/107 (67%), Gaps = 0/107 (0%)
Query 3 INKARRAGDAVETEKKFLGGQNRTTKANLIPNAKKVEEDTGDYHIDRVSTDFSRALAEAR 62
+N ARR+G +ET +KF G N+ + N KK+++DT + DRV T+ +A+ +AR
Sbjct 32 VNAARRSGADIETVRKFNAGSNKAASSGTSLNTKKLDDDTENLSHDRVPTELKKAIMQAR 91
Query 63 RNKGMTQAQLAQAINEKPSVVSEYESGKAIPNGVILQKMSRALGVQL 109
K +TQ+QLA INEKP V+ EYESGKAIPN IL K+ RALG +L
Sbjct 92 GEKKLTQSQLAHLINEKPQVIQEYESGKAIPNQQILSKLERALGAKL 138
> At2g42680
Length=142
Score = 105 bits (261), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 52/107 (48%), Positives = 73/107 (68%), Gaps = 0/107 (0%)
Query 3 INKARRAGDAVETEKKFLGGQNRTTKANLIPNAKKVEEDTGDYHIDRVSTDFSRALAEAR 62
+N ARR+G +ET +KF G N+ + N K +++DT + +RV T+ +A+ +AR
Sbjct 32 VNAARRSGADIETVRKFNAGTNKAASSGTSLNTKMLDDDTENLTHERVPTELKKAIMQAR 91
Query 63 RNKGMTQAQLAQAINEKPSVVSEYESGKAIPNGVILQKMSRALGVQL 109
+K +TQ+QLAQ INEKP V+ EYESGKAIPN IL K+ RALG +L
Sbjct 92 TDKKLTQSQLAQIINEKPQVIQEYESGKAIPNQQILSKLERALGAKL 138
> CE18819
Length=156
Score = 97.8 bits (242), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 51/108 (47%), Positives = 70/108 (64%), Gaps = 2/108 (1%)
Query 2 EINKARRAGDAVETEKKFLGGQNRTTKANLIPNAKKVEEDTGDYHIDRVSTDFSRALAEA 61
++N A+RAG + TEKK + G NR AN N +++E+T + H +V+ + + +A
Sbjct 34 QLNAAQRAGVDISTEKKTMSGGNRQHSAN--KNTLRLDEETEELHHQKVALSLGKVMQQA 91
Query 62 RRNKGMTQAQLAQAINEKPSVVSEYESGKAIPNGVILQKMSRALGVQL 109
R KG TQ L+ INEKP VV EYESGKA+PN I+ KM RALGV+L
Sbjct 92 RATKGWTQKDLSTQINEKPQVVGEYESGKAVPNQQIMAKMERALGVKL 139
> Hs4503453
Length=148
Score = 96.3 bits (238), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 47/109 (43%), Positives = 75/109 (68%), Gaps = 2/109 (1%)
Query 1 QEINKARRAGDAVETEKKFLGGQNRTTKANLIPNAKKVEEDTGDYHIDRVSTDFSRALAE 60
Q I A+R G+ VET KK+ GQN+ + ++ N K++ +T + H DRV+ + + + +
Sbjct 26 QAILAAQRRGEDVETSKKWAAGQNK--QHSITKNTAKLDRETEELHHDRVTLEVGKVIQQ 83
Query 61 ARRNKGMTQAQLAQAINEKPSVVSEYESGKAIPNGVILQKMSRALGVQL 109
R++KG+TQ LA INEKP V+++YESG+AIPN +L K+ RA+G++L
Sbjct 84 GRQSKGLTQKDLATKINEKPQVIADYESGRAIPNNQVLGKIERAIGLKL 132
> At3g24500
Length=148
Score = 94.0 bits (232), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 51/109 (46%), Positives = 70/109 (64%), Gaps = 2/109 (1%)
Query 3 INKARRAGDAVETEKKFLGGQNRTTKANLIP--NAKKVEEDTGDYHIDRVSTDFSRALAE 60
+N A R G AV+T KKF G N+ K+ +P N KK+EE+T +DRV + + +
Sbjct 34 VNAALRNGVAVQTVKKFDAGSNKKGKSTAVPVINTKKLEEETEPAAMDRVKAEVRLMIQK 93
Query 61 ARRNKGMTQAQLAQAINEKPSVVSEYESGKAIPNGVILQKMSRALGVQL 109
AR K M+QA LA+ INE+ VV EYE+GKA+PN +L KM + LGV+L
Sbjct 94 ARLEKKMSQADLAKQINERTQVVQEYENGKAVPNQAVLAKMEKVLGVKL 142
> 7294095
Length=145
Score = 84.3 bits (207), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 44/110 (40%), Positives = 71/110 (64%), Gaps = 8/110 (7%)
Query 3 INKARRAGDAVETEKKFLGGQNR---TTKANLIPNAKKVEEDTGDYHIDRVSTDFSRALA 59
+N+ARR G AV+T++K+ G N+ TTK N K++ +T + D++ D + +
Sbjct 26 VNQARRQGVAVDTQQKYGAGTNKQHVTTK-----NTAKLDRETEELRHDKIPLDVGKLIQ 80
Query 60 EARRNKGMTQAQLAQAINEKPSVVSEYESGKAIPNGVILQKMSRALGVQL 109
+ R++KG++Q LA I EK VV++YE+G+ IPN +IL KM R LG++L
Sbjct 81 QGRQSKGLSQKDLATKICEKQQVVTDYEAGRGIPNNLILGKMERVLGIKL 130
> SPBC83.17
Length=148
Score = 77.8 bits (190), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 45/108 (41%), Positives = 63/108 (58%), Gaps = 2/108 (1%)
Query 2 EINKARRAGDAVETEKKFLGGQNRTTKANLIPNAKKVEEDTGDYHIDRVSTDFSRALAEA 61
+IN ARRAG V TEKK+ G A + K++ + ++A+ +
Sbjct 28 QINSARRAGAIVGTEKKYATGNKSQDPAGQ--HLTKIDRENEVKPPSTTGRSVAQAIQKG 85
Query 62 RRNKGMTQAQLAQAINEKPSVVSEYESGKAIPNGVILQKMSRALGVQL 109
R+ KG Q L+Q INEKP VV++YESG+AIPN +L KM RALG++L
Sbjct 86 RQAKGWAQKDLSQRINEKPQVVNDYESGRAIPNQQVLSKMERALGIKL 133
> YOR298c-a
Length=58
Score = 55.5 bits (132), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 25/43 (58%), Positives = 34/43 (79%), Gaps = 0/43 (0%)
Query 67 MTQAQLAQAINEKPSVVSEYESGKAIPNGVILQKMSRALGVQL 109
M+Q LA INEKP+VV++YE+ +AIPN +L K+ RALGV+L
Sbjct 1 MSQKDLATKINEKPTVVNDYEAARAIPNQQVLSKLERALGVKL 43
> ECU01g0955
Length=95
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 39/71 (54%), Gaps = 3/71 (4%)
Query 38 VEEDTGDYHIDRVSTDFSRALAEARRNKGMTQAQLAQAINEKPSVVSEYESGKAIPNGVI 97
V EDTG + VS A+A AR KGM++ LAQ + + S++ +E G+A+ N I
Sbjct 24 VPEDTG---LRIVSKRVGDAIANARAQKGMSRKDLAQKMKKNVSIIDSWERGEAVYNEKI 80
Query 98 LQKMSRALGVQ 108
++ L V+
Sbjct 81 AKEFESILEVK 91
> At5g14840_3
Length=142
Score = 28.5 bits (62), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 19/66 (28%), Positives = 33/66 (50%), Gaps = 5/66 (7%)
Query 16 EKKFLGGQNRTTKANLIPNAKKVEEDTGDYHIDRVSTDFSRALAEARRNKGMTQAQLAQA 75
E+ + N T +L PN + E+ +G+ H++ + D + + A + +T A A
Sbjct 67 EEHPITSLNIPTSLSLAPNQDQEEKSSGERHLENLDDDTTSSAA-----RTITSAIPQTA 121
Query 76 INEKPS 81
INE PS
Sbjct 122 INEIPS 127
> SPAC8C9.12c
Length=303
Score = 28.5 bits (62), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 11/39 (28%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 11 DAVETEKKFLGGQNRTTKANLIPNAKKVEEDTGDYHIDR 49
DA++T + L G +R+ N++ + K+ G Y + R
Sbjct 41 DAIKTRMQMLNGVSRSVSGNIVNSVIKISSTEGVYSLWR 79
> At3g32917
Length=757
Score = 28.1 bits (61), Expect = 4.1, Method: Composition-based stats.
Identities = 32/100 (32%), Positives = 43/100 (43%), Gaps = 20/100 (20%)
Query 27 TKANLIPNAKKVEEDTGDYHIDRVSTDFSRALAEARRNKGMTQAQLAQAINEKPSV---- 82
TKA L+ A KVEED + R SRA+ + TQ Q+A + KP+
Sbjct 276 TKAALVETAVKVEED-----LQRQVVAVSRAVQPKK-----TQQQVAPSKGNKPAQGQKR 325
Query 83 ----VSEYESGKAIPNGVILQKMSRALGVQLPKCKQKKRV 118
S G PN LQ+M A+ V P +Q+ V
Sbjct 326 KWDHPSRAGQGHLRPNCPKLQRM--AVAVVQPAVQQRAPV 363
> Hs20479818
Length=514
Score = 28.1 bits (61), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 18/68 (26%), Positives = 33/68 (48%), Gaps = 7/68 (10%)
Query 20 LGGQNRTTKANLIPNAKKVEEDTGDYHIDRVSTDFSRALAEARRNKGMTQAQLAQAINEK 79
L +N K++LI +D + +++ TDF A+ + R++ M QA I
Sbjct 239 LKLENIMVKSSLI-------DDNNEINLNIKVTDFGLAVKKQSRSEAMLQATCGTPIYMA 291
Query 80 PSVVSEYE 87
P V+S ++
Sbjct 292 PEVISAHD 299
> At2g32610
Length=748
Score = 26.9 bits (58), Expect = 8.3, Method: Composition-based stats.
Identities = 23/77 (29%), Positives = 35/77 (45%), Gaps = 7/77 (9%)
Query 36 KKVEEDTGDYHIDRVSTDFSRALAEARRNKGMTQAQLAQAINEKPSVVSEYESGKAIPNG 95
+KVE+ TGD H+ V DF A + + N T L + + E V + K IP+
Sbjct 203 RKVEDATGDSHMLDVEDDF-EAFSNTKPNDHST---LVKVVWENKGGVGDE---KEIPHI 255
Query 96 VILQKMSRALGVQLPKC 112
+ + + R V KC
Sbjct 256 IYISREKRPNYVHNQKC 272
Lambda K H
0.310 0.126 0.336
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1198419392
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40