bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
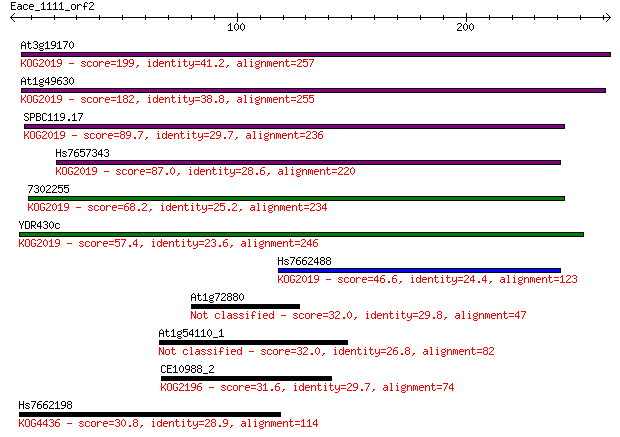
Query= Eace_1111_orf2
Length=262
Score E
Sequences producing significant alignments: (Bits) Value
At3g19170 199 4e-51
At1g49630 182 8e-46
SPBC119.17 89.7 6e-18
Hs7657343 87.0 4e-17
7302255 68.2 2e-11
YDR430c 57.4 3e-08
Hs7662488 46.6 6e-05
At1g72880 32.0 1.3
At1g54110_1 32.0 1.3
CE10988_2 31.6 1.9
Hs7662198 30.8 2.6
> At3g19170
Length=1052
Score = 199 bits (507), Expect = 4e-51, Method: Compositional matrix adjust.
Identities = 106/257 (41%), Positives = 164/257 (63%), Gaps = 5/257 (1%)
Query 6 RMALQLLSYLLLGTSSSPLYKALSESGLGKSVIGGGLGLDLRHATFSVGLKGVAQEEDMV 65
++AL L +L+LGT +SPL K L ESGLG++++ GL +L F +GLKGV++E V
Sbjct 373 QLALGFLDHLMLGTPASPLRKILLESGLGEALVSSGLSDELLQPQFGIGLKGVSEEN--V 430
Query 66 EKVEKVIFRCLSSLSEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSIILDMATESNYD 125
+KVE++I L L+EEGF DA+EA++NT++F+LRE NTG+FP+GLS++L ++ YD
Sbjct 431 QKVEELIMDTLKKLAEEGFDNDAVEASMNTIEFSLRENNTGSFPRGLSLMLQSISKWIYD 490
Query 126 RDPVACLSFDASLKAVREKLKKEDEEGGYSLFQSLIRRFLLNNTHRVTVRLRADPEMTAR 185
DP L + LKA++ ++ EEG ++F LI + +LNN+HRVT+ ++ DPE +
Sbjct 491 MDPFEPLKYTEPLKALKTRIA---EEGSKAVFSPLIEKLILNNSHRVTIEMQPDPEKATQ 547
Query 186 EAKQEKDNLSQIQKQLTPEVVESVLADQIALKQRQLTEDSEEALKTLPVLKLEDVADANN 245
E +EK+ L +++ +T E + + LK +Q T D EAL+ +P L L D+
Sbjct 548 EEVEEKNILEKVKAAMTEEDLAELARATEELKLKQETPDPPEALRCVPSLNLGDIPKEPT 607
Query 246 EVPYEVSALNDVPLISH 262
VP EV +N V ++ H
Sbjct 608 YVPTEVGDINGVKVLRH 624
> At1g49630
Length=1076
Score = 182 bits (461), Expect = 8e-46, Method: Compositional matrix adjust.
Identities = 99/255 (38%), Positives = 160/255 (62%), Gaps = 5/255 (1%)
Query 6 RMALQLLSYLLLGTSSSPLYKALSESGLGKSVIGGGLGLDLRHATFSVGLKGVAQEEDMV 65
++AL L +L+LGT +SPL K L ESGLG++++ G+ +L FS+GLKGV+ +D V
Sbjct 396 QLALGFLDHLMLGTPASPLRKILLESGLGEALVNSGMEDELLQPQFSIGLKGVS--DDNV 453
Query 66 EKVEKVIFRCLSSLSEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSIILDMATESNYD 125
+KVE+++ L L++EGF DA+EA++NT++F+LRE NTG+ P+GLS++L + YD
Sbjct 454 QKVEELVMNTLRKLADEGFDTDAVEASMNTIEFSLRENNTGSSPRGLSLMLQSIAKWIYD 513
Query 126 RDPVACLSFDASLKAVREKLKKEDEEGGYSLFQSLIRRFLLNNTHRVTVRLRADPEMTAR 185
DP L ++ LK+++ ++ E+G S+F LI ++LNN H VT+ ++ DPE +
Sbjct 514 MDPFEPLKYEEPLKSLKARIA---EKGSKSVFSPLIEEYILNNPHCVTIEMQPDPEKASL 570
Query 186 EAKQEKDNLSQIQKQLTPEVVESVLADQIALKQRQLTEDSEEALKTLPVLKLEDVADANN 245
E +EK L +++ +T E + + L+ +Q T D +ALK +P L L D+
Sbjct 571 EEAEEKSILEKVKASMTEEDLTELARATEELRLKQETPDPPDALKCVPSLNLSDIPKEPI 630
Query 246 EVPYEVSALNDVPLI 260
VP EV +N V ++
Sbjct 631 YVPTEVGDINGVKVL 645
> SPBC119.17
Length=882
Score = 89.7 bits (221), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 70/237 (29%), Positives = 113/237 (47%), Gaps = 12/237 (5%)
Query 7 MALQLLSYLLLGTSSSPLYKALSESGLGKSVI-GGGLGLDLRHATFSVGLKGVAQEEDMV 65
AL++LS L SSP YKAL ESGLG G + FSVGL+G ++E +
Sbjct 329 FALKVLSKLCFDGFSSPFYKALIESGLGTDFAPNSGYDSTTKRGIFSVGLEGASEES--L 386
Query 66 EKVEKVIFRCLSSLSEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSIILDMATESNYD 125
K+E +++ + L+ +GF + +EA L+ M+ +L+ + F GL+ L +
Sbjct 387 AKIENLVYSIFNDLALKGFENEKLEAILHQMEISLKH-KSAHFGIGLAQSLPFNWFNG-- 443
Query 126 RDPVACLSFDASLKAVREKLKKEDEEGGYSLFQSLIRRFLLNNTHRVTVRLRADPEMTAR 185
DP LSF+ + E LK+++ +G LFQ LI++++L N R + R
Sbjct 444 ADPADWLSFNKQI----EWLKQKNSDG--KLFQKLIKKYILENKSRFVFTMLPSSTFPQR 497
Query 186 EAKQEKDNLSQIQKQLTPEVVESVLADQIALKQRQLTEDSEEALKTLPVLKLEDVAD 242
+ E L + +LT E + + + L + Q T L TL V + + D
Sbjct 498 LQEAEAKKLQERTSKLTDEDIAEIEKTSVKLLEAQSTPADTSCLPTLSVSDIPETID 554
> Hs7657343
Length=1038
Score = 87.0 bits (214), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 63/221 (28%), Positives = 109/221 (49%), Gaps = 14/221 (6%)
Query 21 SSPLYKALSESGLGKSVIGG-GLGLDLRHATFSVGLKGVAQEEDMVEKVEKVIFRCLSSL 79
+SP YKAL ESGLG G R A FSVGL+G+ +++ +E V +I R + +
Sbjct 358 NSPFYKALIESGLGTEFSPDVGYNGYTREAYFSVGLQGIVEKD--IETVRSLIDRTIDEV 415
Query 80 SEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSIILDMATESNYDRDPVACLSFDASLK 139
E D IEA L+ ++ ++ +T GL + +A+ N+D DPV L L
Sbjct 416 VETRIEDDRIEALLHKIEIQMKHQSTSF---GLMLTSYIASCWNHDGDPVELLKLGNQLA 472
Query 140 AVREKLKKEDEEGGYSLFQSLIRRFLLNNTHRVTVRLRADPEMTAREAKQEKDNLSQIQK 199
R+ L++ + Q ++++ NN H++T+ +R D + ++A+ E L Q +
Sbjct 473 KFRQCLQENPK-----FLQEKVKQYFKNNQHKLTLSMRPDDKYHEKQAQVEATKLKQKVE 527
Query 200 QLTPEVVESVLADQIALKQRQLTEDSEEALKTLPVLKLEDV 240
L+P + + + L+ +Q + LP LK+ D+
Sbjct 528 ALSPGDRQQIYEKGLELRSQQ---SKPQDASCLPALKVSDI 565
> 7302255
Length=1112
Score = 68.2 bits (165), Expect = 2e-11, Method: Composition-based stats.
Identities = 59/237 (24%), Positives = 105/237 (44%), Gaps = 18/237 (7%)
Query 9 LQLLSYLLLGTSSSPLYKALSESGL-GKSVIGGGLGLDLRHATFSVGLKGVAQEE--DMV 65
L +LS +L+ +SP YK L E G G D + TF VGL+ + E+ +
Sbjct 371 LHVLSEVLIRGPNSPFYKNLIEPNFSGGYNQTTGYSSDTKDTTFVVGLQDLRVEDFKKCI 430
Query 66 EKVEKVIFRCLSSLSEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSIILDMATESNYD 125
E +K I ++ +GF +E+ L+ ++ +L+ N G +++ + N+D
Sbjct 431 EIFDKTIINSMN----DGFDSQHVESVLHNLELSLKHQNPNF---GNTLLFNSTALWNHD 483
Query 126 RDPVACLSFDASLKAVREKLKKEDEEGGYSLFQSLIRRFLLNNTHRVTVRLRADPEMTAR 185
D V+ L + +RE + + + FQ I ++ NN HR+T+ + D +
Sbjct 484 GDVVSNLRVSDMISGLRESISQNKK-----YFQEKIEKYFANNNHRLTLTMSPDEAYEDK 538
Query 186 EAKQEKDNLSQIQKQLTPEVVESVLADQIALKQRQLTEDSEEALKTLPVLKLEDVAD 242
+ E + + Q K L +E + + L Q E + + LP L + DV D
Sbjct 539 FKQAELELVEQKVKLLDEVKIEKIYERGLILDSYQKAESNTD---LLPCLTMNDVRD 592
> YDR430c
Length=989
Score = 57.4 bits (137), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 58/247 (23%), Positives = 114/247 (46%), Gaps = 14/247 (5%)
Query 5 DRMALQLLSYLLLGTSSSPLYKALSESGLG-KSVIGGGLGLDLRHATFSVGLKGVAQEED 63
D L++L LL+ SS +Y+ L ESG+G + + G+ +VG++GV+ E
Sbjct 317 DTFLLKVLGNLLMDGHSSVMYQKLIESGIGLEFSVNSGVEPTTAVNLLTVGIQGVSDIEI 376
Query 64 MVEKVEKVIFRCLSSLSEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSIILDMATESN 123
+ V + L + E F + I+A + ++ + ++ GL ++ +
Sbjct 377 FKDTVNNIFQNLLET--EHPFDRKRIDAIIEQLELSKKDQKADF---GLQLLYSILPGWT 431
Query 124 YDRDPVACLSFDASLKAVREKLKKEDEEGGYSLFQSLIRRFLLNNTHRVTVRLRADPEMT 183
DP L F+ L+ R L E G +LFQ LIR+++++ T ++ E +
Sbjct 432 NKIDPFESLLFEDVLQRFRGDL----ETKGDTLFQDLIRKYIVHKP-CFTFSIQGSEEFS 486
Query 184 AREAKQEKDNLSQIQKQLTPEVVESVLADQIALKQRQLTEDSEEALKTLPVLKLEDVADA 243
+E+ L + L + +++ I L+++Q + +E L LP L+++D+ A
Sbjct 487 KSLDDEEQTRLREKITALDEQDKKNIFKRGILLQEKQ---NEKEDLSCLPTLQIKDIPRA 543
Query 244 NNEVPYE 250
++ E
Sbjct 544 GDKYSIE 550
> Hs7662488
Length=595
Score = 46.6 bits (109), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 30/123 (24%), Positives = 59/123 (47%), Gaps = 8/123 (6%)
Query 118 MATESNYDRDPVACLSFDASLKAVREKLKKEDEEGGYSLFQSLIRRFLLNNTHRVTVRLR 177
+A+ N+D DPV L L R+ L++ + Q ++++ NN H++T+ +R
Sbjct 9 IASCWNHDGDPVELLKLGNQLAKFRQCLQENPK-----FLQEKVKQYFKNNQHKLTLSMR 63
Query 178 ADPEMTAREAKQEKDNLSQIQKQLTPEVVESVLADQIALKQRQLTEDSEEALKTLPVLKL 237
D + ++A+ E L Q + L+P + + + L+ +Q + LP LK+
Sbjct 64 PDDKYHEKQAQVEATKLKQKVEALSPGDRQQIYEKGLELRSQQ---SKPQDASCLPALKV 120
Query 238 EDV 240
D+
Sbjct 121 SDI 123
> At1g72880
Length=385
Score = 32.0 bits (71), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 27/47 (57%), Gaps = 0/47 (0%)
Query 80 SEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSIILDMATESNYDR 126
S+E KDA+ L + T+R++ G FPK S+ +++ T + ++
Sbjct 198 SQESHFKDAVGVCLPLINATIRDIAKGVFPKDCSLNIEIPTSPSSNK 244
> At1g54110_1
Length=198
Score = 32.0 bits (71), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 22/87 (25%), Positives = 42/87 (48%), Gaps = 7/87 (8%)
Query 66 EKVEKVIFRCL-----SSLSEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSIILDMAT 120
EK+E V+ R + + +S+E F+KD+ + ++ R N+ + I+ +
Sbjct 62 EKIEAVVSRIVAQVPHTEVSDEAFAKDSTNDSSPKVEDDTRTPNSPQLRR--RIVPASSK 119
Query 121 ESNYDRDPVACLSFDASLKAVREKLKK 147
E +YD DP + D + +A K +K
Sbjct 120 EQSYDADPSKPIKLDTAAQAQVNKQRK 146
> CE10988_2
Length=197
Score = 31.6 bits (70), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 22/82 (26%), Positives = 39/82 (47%), Gaps = 8/82 (9%)
Query 67 KVEKVIFRCLSSLSEEGFSKDAIEAALNTMQFTLRELNTGT--------FPKGLSIILDM 118
++ +++ S + F D I +LNTMQ T+ + T T K L ++D+
Sbjct 110 QIAQMMLNVDSQMKCADFDLDQITKSLNTMQSTVLKTKTETPLEKTELIMKKQLQKLMDL 169
Query 119 ATESNYDRDPVACLSFDASLKA 140
+T+ + RD + L D +LK
Sbjct 170 STQHDATRDKLNKLKDDHNLKT 191
> Hs7662198
Length=1299
Score = 30.8 bits (68), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 33/122 (27%), Positives = 55/122 (45%), Gaps = 10/122 (8%)
Query 5 DRMALQLLSY---LLLGTSSSPLYKALSESGLGKSVIGGGLGLDLRHATFSVGLKGVAQE 61
D M+LQ+ Y LL LY L E+ + S+ L L + FS+G VA+
Sbjct 1048 DMMSLQIQMYQLSRLLHDYHRDLYNHLEENEISPSLYAAPWFLTLFASQFSLGF--VARV 1105
Query 62 EDMV-----EKVEKVIFRCLSSLSEEGFSKDAIEAALNTMQFTLRELNTGTFPKGLSIIL 116
D++ E + KV LSS ++ E + ++ TL ++NT K ++ +
Sbjct 1106 FDIIFLQGTEVIFKVALSLLSSQETLIMECESFENIVEFLKNTLPDMNTSEMEKIITQVF 1165
Query 117 DM 118
+M
Sbjct 1166 EM 1167
Lambda K H
0.313 0.131 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5542830696
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40