bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1101_orf2
Length=239
Score E
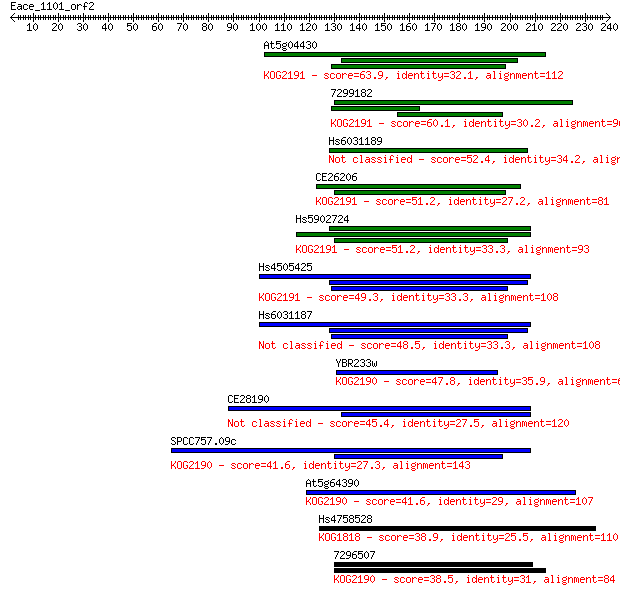
Sequences producing significant alignments: (Bits) Value
At5g04430 63.9 3e-10
7299182 60.1 4e-09
Hs6031189 52.4 8e-07
CE26206 51.2 2e-06
Hs5902724 51.2 2e-06
Hs4505425 49.3 7e-06
Hs6031187 48.5 1e-05
YBR233w 47.8 2e-05
CE28190 45.4 1e-04
SPCC757.09c 41.6 0.002
At5g64390 41.6 0.002
Hs4758528 38.9 0.010
7296507 38.5 0.014
> At5g04430
Length=313
Score = 63.9 bits (154), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 36/114 (31%), Positives = 66/114 (57%), Gaps = 3/114 (2%)
Query 102 GAPTANSATAAAATAATAAAAAADTCYCKLLVSNHLAGIIIGQAGSEIRTLKTLTGAKIV 161
G+P + + ++ A +A + + LVSN AG +IG+ GS I + +GA+I
Sbjct 10 GSPEELAKRSPEPHDSSEADSAEKPTHIRFLVSNAAAGSVIGKGGSTITEFQAKSGARIQ 69
Query 162 LSPHGMYFPGTSDRVAAIEGAETAVLHVVDWLIDKLQTA--AEDAANPLQQQQQ 213
LS + +FPGT+DR+ I G+ V++ ++ ++DKL + AED N ++ +++
Sbjct 70 LSRNQEFFPGTTDRIIMISGSIKEVVNGLELILDKLHSELHAED-GNEVEPRRR 122
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 39/70 (55%), Gaps = 0/70 (0%)
Query 133 VSNHLAGIIIGQAGSEIRTLKTLTGAKIVLSPHGMYFPGTSDRVAAIEGAETAVLHVVDW 192
V++ G+++G+ G I + +TGA+I +S G + GT+DR +I G + A+
Sbjct 240 VADEHIGLVLGRGGRNIMEITQMTGARIKISDRGDFMSGTTDRKVSITGPQRAIQQAETM 299
Query 193 LIDKLQTAAE 202
+ K+ +A E
Sbjct 300 IKQKVDSATE 309
Score = 40.4 bits (93), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 36/69 (52%), Gaps = 0/69 (0%)
Query 129 CKLLVSNHLAGIIIGQAGSEIRTLKTLTGAKIVLSPHGMYFPGTSDRVAAIEGAETAVLH 188
+L+V N G IIG+ G+ I++ + A I +SP F G SDR+ + G +
Sbjct 123 IRLVVPNSSCGGIIGKGGATIKSFIEESKAGIKISPLDNTFYGLSDRLVTLSGTFEEQMR 182
Query 189 VVDWLIDKL 197
+D ++ KL
Sbjct 183 AIDLILAKL 191
> 7299182
Length=493
Score = 60.1 bits (144), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 29/95 (30%), Positives = 56/95 (58%), Gaps = 0/95 (0%)
Query 130 KLLVSNHLAGIIIGQAGSEIRTLKTLTGAKIVLSPHGMYFPGTSDRVAAIEGAETAVLHV 189
K+LV +G IIG+ G I +L+ TGA++ +S ++PGT++RV I G+ A++ V
Sbjct 2 KILVPAVASGAIIGKGGETIASLQKDTGARVKMSKSHDFYPGTTERVCLITGSTEAIMVV 61
Query 190 VDWLIDKLQTAAEDAANPLQQQQQQQQQMQMQQQI 224
+++++DK++ + + +Q Q+ Q +I
Sbjct 62 MEFIMDKIREKPDLTNKIVDTDSKQTQERDKQVKI 96
Score = 31.6 bits (70), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 24/35 (68%), Gaps = 0/35 (0%)
Query 129 CKLLVSNHLAGIIIGQAGSEIRTLKTLTGAKIVLS 163
K+LV N AG+IIG+ G+ I+ +K +G+ + +S
Sbjct 94 VKILVPNSTAGMIIGKGGAFIKQIKEESGSYVQIS 128
Score = 30.0 bits (66), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 13/42 (30%), Positives = 26/42 (61%), Gaps = 1/42 (2%)
Query 155 LTGAKIVLSPHGMYFPGTSDRVAAIEGAETAVLHVVDWLIDK 196
++GA + +S G++ PGT +R+ I G +A+ +LI++
Sbjct 433 VSGANVQISKKGIFAPGTRNRIVTITGQPSAIAK-AQYLIEQ 473
> Hs6031189
Length=181
Score = 52.4 bits (124), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 47/79 (59%), Gaps = 0/79 (0%)
Query 128 YCKLLVSNHLAGIIIGQAGSEIRTLKTLTGAKIVLSPHGMYFPGTSDRVAAIEGAETAVL 187
+ K+L+ ++ AG IIG+ G I L+ TGA I LS ++PGT++RV I+G A+
Sbjct 51 FLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKSKDFYPGTTERVCLIQGTVEALN 110
Query 188 HVVDWLIDKLQTAAEDAAN 206
V ++ +K++ ++ A
Sbjct 111 AVHGFIAEKIREMPQNVAK 129
> CE26206
Length=413
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 22/81 (27%), Positives = 44/81 (54%), Gaps = 0/81 (0%)
Query 123 AADTCYCKLLVSNHLAGIIIGQAGSEIRTLKTLTGAKIVLSPHGMYFPGTSDRVAAIEGA 182
D K+L+ ++ G IIG+ G +R LK ++ +S + +PGTS+R+ ++G
Sbjct 38 GGDHLSIKILIPSNAVGAIIGKGGEAMRNLKNDNNCRVQMSKNSETYPGTSERICLVKGR 97
Query 183 ETAVLHVVDWLIDKLQTAAED 203
++ V++ + DK++ D
Sbjct 98 LNNIMAVIESIQDKIREKCAD 118
Score = 29.3 bits (64), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 41/71 (57%), Gaps = 3/71 (4%)
Query 130 KLLVSNHLAGIIIGQAGSEIRTLKTLTGAKIVLSPH-GMYFPGTS-DRVAAIEGAE-TAV 186
K+++ N AG++IG++G+ I+ ++ G +I + P G TS +RV + E +A+
Sbjct 137 KIVMPNTSAGMVIGKSGANIKDIREQFGCQIQVYPKAGSVEAKTSLERVVTVAHDEASAL 196
Query 187 LHVVDWLIDKL 197
L +++K+
Sbjct 197 LQAASRVLEKV 207
> Hs5902724
Length=492
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 26/80 (32%), Positives = 46/80 (57%), Gaps = 0/80 (0%)
Query 128 YCKLLVSNHLAGIIIGQAGSEIRTLKTLTGAKIVLSPHGMYFPGTSDRVAAIEGAETAVL 187
+ K+L+ ++ AG IIG+ G I L+ TGA I LS ++PGT++RV ++G A+
Sbjct 34 FLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKSKDFYPGTTERVCLVQGTAEALN 93
Query 188 HVVDWLIDKLQTAAEDAANP 207
V ++ +K++ + P
Sbjct 94 AVHSFIAEKVREIPQAMTKP 113
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 31/95 (32%), Positives = 48/95 (50%), Gaps = 4/95 (4%)
Query 115 TAATAAAAAADTCYCKLLVSNHLAGIIIGQAGSEIRTLKTLTGAKIVLSPHGMYFPGTSD 174
TA AA +A ++ V +L G I+G+ G + + LTGA+I +S G + PGT +
Sbjct 396 TAEKLAAESAKEL-VEIAVPENLVGAILGKGGKTLVEYQELTGARIQISKKGEFLPGTRN 454
Query 175 RVAAIEGAETAVLHVVDWLIDKLQTAAE--DAANP 207
R I G+ A +LI + T + A+NP
Sbjct 455 RRVTITGSPAAT-QAAQYLISQRVTYEQGVRASNP 488
Score = 39.3 bits (90), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 38/71 (53%), Gaps = 5/71 (7%)
Query 130 KLLVSNHLAGIIIGQAGSEIRTLKTLTGAKIVLS--PHGMYFPGTSDRVAAIEGAETAVL 187
KL+V N AG+IIG+ G+ ++ + +GA + LS P G+ +RV + G V
Sbjct 134 KLIVPNSTAGLIIGKGGATVKAVMEQSGAWVQLSQKPEGINL---QERVVTVSGEPEQVH 190
Query 188 HVVDWLIDKLQ 198
V ++ K+Q
Sbjct 191 KAVSAIVQKVQ 201
> Hs4505425
Length=510
Score = 49.3 bits (116), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 36/110 (32%), Positives = 52/110 (47%), Gaps = 6/110 (5%)
Query 100 YFGAPTANSATAAAATAATAAAAAADTCYCKLLVSNHLAGIIIGQAGSEIRTLKTLTGAK 159
YFGA S AA+A T + ++ V +L G I+G+ G + + LTGA+
Sbjct 401 YFGA---ASPLAASAILGTEKSTDGSKDVVEIAVPENLVGAILGKGGKTLVEYQELTGAR 457
Query 160 IVLSPHGMYFPGTSDRVAAIEGAETAVLHVVDWLIDKLQTAAE--DAANP 207
I +S G + PGT +R I G A +LI + T + AANP
Sbjct 458 IQISKKGEFVPGTRNRKVTITGTPAAT-QAAQYLITQRITYEQGVRAANP 506
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 47/82 (57%), Gaps = 3/82 (3%)
Query 128 YCKLLVSNHLAGIIIGQAGSEIRTLKTLTGAKIVLSPHGM---YFPGTSDRVAAIEGAET 184
+ K+L+ ++ AG IIG+ G I L+ TGA I LS ++PGT++RV I+G
Sbjct 51 FLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKLSKSKDFYPGTTERVCLIQGTVE 110
Query 185 AVLHVVDWLIDKLQTAAEDAAN 206
A+ V ++ +K++ ++ A
Sbjct 111 ALNAVHGFIAEKIREMPQNVAK 132
Score = 39.3 bits (90), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 38/72 (52%), Gaps = 5/72 (6%)
Query 129 CKLLVSNHLAGIIIGQAGSEIRTLKTLTGAKIVLS--PHGMYFPGTSDRVAAIEGAETAV 186
K++V N AG+IIG+ G+ ++ + +GA + LS P G+ +RV + G
Sbjct 177 VKIIVPNSTAGLIIGKGGATVKAVMEQSGAWVQLSQKPDGINL---QERVVTVSGEPEQN 233
Query 187 LHVVDWLIDKLQ 198
V+ +I K+Q
Sbjct 234 RKAVELIIQKIQ 245
> Hs6031187
Length=486
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 36/110 (32%), Positives = 52/110 (47%), Gaps = 6/110 (5%)
Query 100 YFGAPTANSATAAAATAATAAAAAADTCYCKLLVSNHLAGIIIGQAGSEIRTLKTLTGAK 159
YFGA S AA+A T + ++ V +L G I+G+ G + + LTGA+
Sbjct 377 YFGA---ASPLAASAILGTEKSTDGSKDVVEIAVPENLVGAILGKGGKTLVEYQELTGAR 433
Query 160 IVLSPHGMYFPGTSDRVAAIEGAETAVLHVVDWLIDKLQTAAE--DAANP 207
I +S G + PGT +R I G A +LI + T + AANP
Sbjct 434 IQISKKGEFVPGTRNRKVTITGTPAAT-QAAQYLITQRITYEQGVRAANP 482
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 47/82 (57%), Gaps = 3/82 (3%)
Query 128 YCKLLVSNHLAGIIIGQAGSEIRTLKTLTGAKIVLSPHGM---YFPGTSDRVAAIEGAET 184
+ K+L+ ++ AG IIG+ G I L+ TGA I LS ++PGT++RV I+G
Sbjct 51 FLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKLSKSKDFYPGTTERVCLIQGTVE 110
Query 185 AVLHVVDWLIDKLQTAAEDAAN 206
A+ V ++ +K++ ++ A
Sbjct 111 ALNAVHGFIAEKIREMPQNVAK 132
Score = 38.5 bits (88), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 38/72 (52%), Gaps = 5/72 (6%)
Query 129 CKLLVSNHLAGIIIGQAGSEIRTLKTLTGAKIVLS--PHGMYFPGTSDRVAAIEGAETAV 186
K++V N AG+IIG+ G+ ++ + +GA + LS P G+ +RV + G
Sbjct 153 VKIIVPNSTAGLIIGKGGATVKAVMEQSGAWVQLSQKPDGINL---QERVVTVSGEPEQN 209
Query 187 LHVVDWLIDKLQ 198
V+ +I K+Q
Sbjct 210 RKAVELIIQKIQ 221
> YBR233w
Length=413
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 39/64 (60%), Gaps = 3/64 (4%)
Query 131 LLVSNHLAGIIIGQAGSEIRTLKTLTGAKIVLSPHGMYFPGTSDRVAAIEGAETAVLHVV 190
LL+ +HL G IIG+ GS +R ++ L+ AK+ SP+ + ++DR+ I G A+ H+
Sbjct 153 LLIPHHLMGCIIGKRGSRLREIEDLSAAKLFASPNQLLL--SNDRILTINGVPDAI-HIA 209
Query 191 DWLI 194
+ I
Sbjct 210 TFYI 213
> CE28190
Length=557
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 33/120 (27%), Positives = 48/120 (40%), Gaps = 7/120 (5%)
Query 88 NTSSNHSSNAPFYFGAPTANSATAAAATAATAAAAAADTCYCKLLVSNHLAGIIIGQAGS 147
N +N NAP AP S A A A + +L AG+IIG+ G
Sbjct 206 NIMNNTQGNAPLLQRAPHQPSGQFGGGYGAQEAQAKGEVIVPRLS-----AGMIIGKGGE 260
Query 148 EIRTLKTLTGAKIVLSPHGMYFPGTSDRVAAIEGAETAVLHVVDWLIDKLQTAAEDAANP 207
I+ L TG KI P P + DR+A I G + + + + + A ++ P
Sbjct 261 MIKRLAAETGTKIQFKPD--TNPNSEDRIAVIMGTRDQIYRATERITEIVNRAIKNNGAP 318
Score = 36.2 bits (82), Expect = 0.064, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 39/78 (50%), Gaps = 5/78 (6%)
Query 133 VSNHLAGIIIGQAGSEIRTLKTLTGAKIVLSPHGMYFPGTSDRVAAIEGAETAV---LHV 189
V + G++IG+ GSEIR ++ +G ++ + P G R IEG V +
Sbjct 60 VPEKVVGLVIGKGGSEIRLIQQTSGCRVQMDPDHQSVNGF--RNCTIEGPPDQVAVARQM 117
Query 190 VDWLIDKLQTAAEDAANP 207
+ +I++ QT A+ A P
Sbjct 118 ITQVINRNQTGAQPGAAP 135
> SPCC757.09c
Length=398
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 39/143 (27%), Positives = 59/143 (41%), Gaps = 17/143 (11%)
Query 65 LPSIAAALQTSTRSSNSTICSSSNTSSNHSSNAPFYFGAPTANSATAAAATAATAAAAAA 124
+P+ + ST ++ STI S +S P P + AT A A
Sbjct 43 IPTPKPSTPLSTLTNGSTIQQSMTNQPEPTSQVPPISAKPPMDDATYATQQLTLRA---- 98
Query 125 DTCYCKLLVSNHLAGIIIGQAGSEIRTLKTLTGAKIVLSPHGMYFPGTSDRVAAIEGAET 184
L+S AGIIIG+AG + L++ T K ++ P DRV I G
Sbjct 99 -------LLSTREAGIIIGKAGKNVAELRSTTNVKAGVT---KAVPNVHDRVLTISGPLE 148
Query 185 AVLHVVDWLIDKLQTAAEDAANP 207
V+ ++ID A+++ NP
Sbjct 149 NVVRAYRFIIDIF---AKNSTNP 168
Score = 30.4 bits (67), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 33/67 (49%), Gaps = 3/67 (4%)
Query 130 KLLVSNHLAGIIIGQAGSEIRTLKTLTGAKIVLSPHGMYFPGTSDRVAAIEGAETAVLHV 189
+LL+++ L G IIG+ G I+ ++ +++ S P +++R I G LH
Sbjct 182 RLLIAHSLMGSIIGRNGLRIKLIQDKCSCRMIASKD--MLPQSTERTVEIHGT-VDNLHA 238
Query 190 VDWLIDK 196
W I K
Sbjct 239 AIWEIGK 245
> At5g64390
Length=833
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 31/117 (26%), Positives = 53/117 (45%), Gaps = 13/117 (11%)
Query 119 AAAAAADTCYCKLLVSNHLAGIIIGQAGSEIRTLKTLTGAKIVLSPHGMYF-PGTSDRVA 177
AA A +DT C+LL + AG +IG+ G + +++ TG KI + + T D +
Sbjct 142 AAEADSDTVVCRLLTESSHAGAVIGKGGQMVGSIRKETGCKISIRIENLPICADTDDEMV 201
Query 178 AIEGAETAVLHVVDWL---------IDKLQTAAEDAANPLQQQQQQQQQMQMQQQIQ 225
+EG AV + + IDK++ PL+++ Q ++ IQ
Sbjct 202 EVEGNAIAVKKALVSISRCLQNCQSIDKVRMVGN---RPLEKEFQASLHRPIETIIQ 255
> Hs4758528
Length=777
Score = 38.9 bits (89), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 28/117 (23%), Positives = 61/117 (52%), Gaps = 15/117 (12%)
Query 124 ADTCYCKLLVSNHLAGIIIGQAGSEIRTLKTLTG--AKIV-----LSPHGMYFPGTSDRV 176
A T + + SNH+ G I + + +++ G +++ L +Y+ G D++
Sbjct 415 AVTTFVNRMKSNHMRGRSITNDSAVLSLFQSINGMHPQLLELLNQLDERRLYYEGLQDKL 474
Query 177 AAIEGAETAVLHVVDWLIDKLQTAAEDAANPLQQQQQQQQQMQMQQQIQLQMQQQQQ 233
A I A A+ + + +KL+ AAE+A ++Q+Q+Q+ Q++++ Q++Q+
Sbjct 475 AQIRDARGALSALREEHREKLRRAAEEA--------ERQRQIQLAQKLEIMRQKKQE 523
> 7296507
Length=386
Score = 38.5 bits (88), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 44/79 (55%), Gaps = 2/79 (2%)
Query 130 KLLVSNHLAGIIIGQAGSEIRTLKTLTGAKIVLSPHGMYFPGTSDRVAAIEGAETAVLHV 189
++ VSN L G IIG+ G++I ++ ++GA I +S G +DR I G +V +
Sbjct 293 EMTVSNDLIGCIIGKGGTKIAEIRQISGAMIRISNCEEREGGNTDRTITISGNPDSV-AL 351
Query 190 VDWLIDKLQTAAEDAANPL 208
+LI+ ++ + E A P+
Sbjct 352 AQYLIN-MRISMETAGLPI 369
Score = 28.9 bits (63), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 20/85 (23%), Positives = 41/85 (48%), Gaps = 3/85 (3%)
Query 130 KLLVSNHLAGIIIGQAGSEIRTLKTLTGAKIVLSPHGMYFPGTSDRVAAIEG-AETAVLH 188
+L+V G +IG++GS+I+ ++ TG I ++ P +++R + G AE
Sbjct 110 RLIVPASQCGSLIGKSGSKIKEIRQTTGCSIQVASE--MLPNSTERAVTLSGSAEQITQC 167
Query 189 VVDWLIDKLQTAAEDAANPLQQQQQ 213
+ + L++ A P + + Q
Sbjct 168 IYQICLVMLESPPRGATIPYRPKPQ 192
Lambda K H
0.309 0.119 0.324
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4776280724
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40