bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
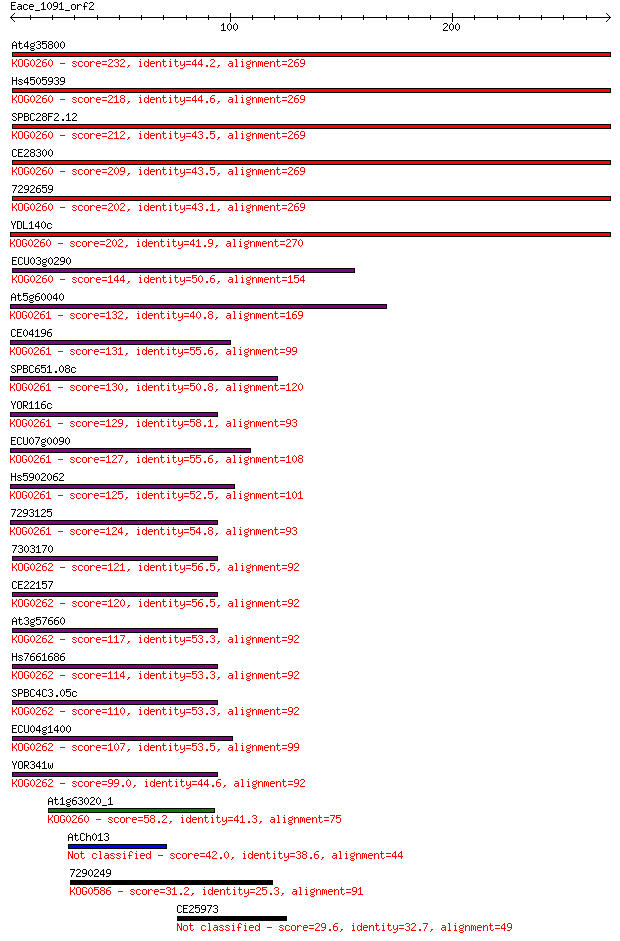
Query= Eace_1091_orf2
Length=270
Score E
Sequences producing significant alignments: (Bits) Value
At4g35800 232 6e-61
Hs4505939 218 8e-57
SPBC28F2.12 212 8e-55
CE28300 209 3e-54
7292659 202 6e-52
YDL140c 202 7e-52
ECU03g0290 144 3e-34
At5g60040 132 5e-31
CE04196 131 2e-30
SPBC651.08c 130 2e-30
YOR116c 129 8e-30
ECU07g0090 127 2e-29
Hs5902062 125 1e-28
7293125 124 2e-28
7303170 121 1e-27
CE22157 120 3e-27
At3g57660 117 2e-26
Hs7661686 114 2e-25
SPBC4C3.05c 110 3e-24
ECU04g1400 107 2e-23
YOR341w 99.0 9e-21
At1g63020_1 58.2 2e-08
AtCh013 42.0 0.002
7290249 31.2 2.1
CE25973 29.6 7.2
> At4g35800
Length=1840
Score = 232 bits (591), Expect = 6e-61, Method: Compositional matrix adjust.
Identities = 119/269 (44%), Positives = 174/269 (64%), Gaps = 5/269 (1%)
Query 2 RSLPHFSKFDYSPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRL 61
R+LPHF+K DY PESRGFVE+SYL GL PQE FFHAMGGREG+IDTA KTSETGYIQRRL
Sbjct 802 RTLPHFTKDDYGPESRGFVENSYLRGLTPQEFFFHAMGGREGLIDTAVKTSETGYIQRRL 861
Query 62 MKAMEDVKVSYDRTVRNSAQEVLQFLYGEDGMSGEYVEDQVIELMQMDAEKIRRVYRHDL 121
+KAMED+ V YD TVRNS +V+QFLYGEDGM ++E Q ++ ++M + R +++++
Sbjct 862 VKAMEDIMVKYDGTVRNSLGDVIQFLYGEDGMDAVWIESQKLDSLKMKKSEFDRTFKYEI 921
Query 122 DQESYGKGWILNEELRTKLFVDFSAQQVLEDEYEAIKEDKLRLCKSIFPDGEVKQHIPIN 181
D E++ ++ +E L + + V + EY ++ D+ +L I +G+ +P+N
Sbjct 922 DDENWNPTYLSDEHLEDLKGIR-ELRDVFDAEYSKLETDRFQLGTEIATNGDSTWPLPVN 980
Query 182 IMRLLEFAKAQFPPSVEQLKKQDPVAIAKSVSELLNDKLIVVKQTCKADTISAEVQENAT 241
I R + A+ F + ++ PV I +V + L ++L+VV D +S E Q+NAT
Sbjct 981 IKRHIWNAQKTFKIDLRKISDMHPVEIVDAVDK-LQERLLVVPGD---DALSVEAQKNAT 1036
Query 242 VFMKAHLRTVLNSRRLMEREQLGVKAVHW 270
+F LR+ L S+R++E +L +A W
Sbjct 1037 LFFNILLRSTLASKRVLEEYKLSREAFEW 1065
> Hs4505939
Length=1970
Score = 218 bits (556), Expect = 8e-57, Method: Compositional matrix adjust.
Identities = 120/269 (44%), Positives = 167/269 (62%), Gaps = 6/269 (2%)
Query 2 RSLPHFSKFDYSPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRL 61
R+LPHF K DY PESRGFVE+SYL GL P E FFHAMGGREG+IDTA KT+ETGYIQRRL
Sbjct 805 RTLPHFIKDDYGPESRGFVENSYLAGLTPTEFFFHAMGGREGLIDTAVKTAETGYIQRRL 864
Query 62 MKAMEDVKVSYDRTVRNSAQEVLQFLYGEDGMSGEYVEDQVIELMQMDAEKIRRVYRHDL 121
+K+ME V V YD TVRNS +V+Q YGEDG++GE VE Q + ++ + + +R D
Sbjct 865 IKSMESVMVKYDATVRNSINQVVQLRYGEDGLAGESVEFQNLATLKPSNKAFEKKFRFDY 924
Query 122 DQESYGKGWILNEELRTKLFVDFSAQQVLEDEYEAIKEDKLRLCKSIFPDGEVKQHIPIN 181
E + L E+L + + Q LE E+E ++ED+ + + IFP G+ K +P N
Sbjct 925 TNERALR-RTLQEDLVKDVLSNAHIQNELEREFERMREDR-EVLRVIFPTGDSKVVLPCN 982
Query 182 IMRLLEFAKAQFPPSVEQLKKQDPVAIAKSVSELLNDKLIVVKQTCKADTISAEVQENAT 241
++R++ A+ F + P+ + + V E L+ KL++V D +S + QENAT
Sbjct 983 LLRMIWNAQKIFHINPRLPSDLHPIKVVEGVKE-LSKKLVIVNGD---DPLSRQAQENAT 1038
Query 242 VFMKAHLRTVLNSRRLMEREQLGVKAVHW 270
+ HLR+ L SRR+ E +L +A W
Sbjct 1039 LLFNIHLRSTLCSRRMAEEFRLSGEAFDW 1067
> SPBC28F2.12
Length=1752
Score = 212 bits (539), Expect = 8e-55, Method: Compositional matrix adjust.
Identities = 117/271 (43%), Positives = 169/271 (62%), Gaps = 13/271 (4%)
Query 2 RSLPHFSKFDYSPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRL 61
R+LPHF K D SPESRGF+E+SYL GL PQE FFHAM GREG+IDTA KT+ETGYIQRRL
Sbjct 788 RTLPHFPKDDDSPESRGFIENSYLRGLTPQEFFFHAMAGREGLIDTAVKTAETGYIQRRL 847
Query 62 MKAMEDVKVSYDRTVRNSAQEVLQFLYGEDGMSGEYVEDQVIELMQMDAEKIRRVYRHDL 121
+KAMEDV V YD TVRN+ +++QF YGEDG+ VE QV + +++ ++ + YR DL
Sbjct 848 VKAMEDVMVRYDGTVRNAMGDIIQFAYGEDGLDATLVEYQVFDSLRLSTKQFEKKYRIDL 907
Query 122 DQESYGKGWILNEELRTKLFVDFSAQQVLEDEYEAIKEDKLRLCKSIFPDGEVKQHIPIN 181
++ L+ + + D S Q +L++EY + D+ LCK IFP G+ + +P+N
Sbjct 908 MEDRS-----LSLYMENSIENDSSVQDLLDEEYTQLVADRELLCKFIFPKGDARWPLPVN 962
Query 182 IMRLLEFAKAQFPPSVEQLKKQD--PVAIAKSVSELLNDKLIVVKQTCKADTISAEVQEN 239
+ R+++ A F +E K D P I ++EL+ KL + + +D I+ +VQ N
Sbjct 963 VQRIIQNALQIF--HLEAKKPTDLLPSDIINGLNELIA-KLTIFR---GSDRITRDVQNN 1016
Query 240 ATVFMKAHLRTVLNSRRLMEREQLGVKAVHW 270
AT+ + LR+ +R++ +L A W
Sbjct 1017 ATLLFQILLRSKFAVKRVIMEYRLNKVAFEW 1047
> CE28300
Length=1852
Score = 209 bits (533), Expect = 3e-54, Method: Compositional matrix adjust.
Identities = 117/270 (43%), Positives = 168/270 (62%), Gaps = 8/270 (2%)
Query 2 RSLPHFSKFDYSPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRL 61
R+LPHF K DY PES+GFVE+SYL GL P E FFHAMGGREG+IDTA KT+ETGYIQRRL
Sbjct 795 RTLPHFIKDDYGPESKGFVENSYLAGLTPSEFFFHAMGGREGLIDTAVKTAETGYIQRRL 854
Query 62 MKAMEDVKVSYDRTVRNSAQEVLQFLYGEDGMSGEYVEDQVIELMQMDAEKIRRVYRHDL 121
+KAME V V+YD TVRNS +++Q YGEDG+ G +VE+Q + M+ + R +R DL
Sbjct 855 IKAMESVMVNYDGTVRNSLAQMVQLRYGEDGLDGMWVENQNMPTMKPNNAVFERDFRMDL 914
Query 122 -DQESYGKGWILNEELRTKLFVDFSAQQVLEDEYEAIKEDKLRLCKSIFPDGEVKQHIPI 180
D + K + +E++ ++ ++E E+ ++ED+ RL + IFP G+ K +P
Sbjct 915 TDNKFLRKNY--SEDVVREIQESEDGISLVESEWSQLEEDR-RLLRKIFPRGDAKIVLPC 971
Query 181 NIMRLLEFAKAQFPPSVEQLKKQDPVAIAKSVSELLNDKLIVVKQTCKADTISAEVQENA 240
N+ RL+ A+ F + + P+ + V E L+ KLI+V D IS + Q NA
Sbjct 972 NLQRLIWNAQKIFKVDLRKPVNLSPLHVISGVRE-LSKKLIIVSGN---DEISKQAQYNA 1027
Query 241 TVFMKAHLRTVLNSRRLMEREQLGVKAVHW 270
T+ M LR+ L ++ + + +L +A W
Sbjct 1028 TLLMNILLRSTLCTKNMCTKSKLNSEAFDW 1057
> 7292659
Length=1887
Score = 202 bits (514), Expect = 6e-52, Method: Compositional matrix adjust.
Identities = 116/278 (41%), Positives = 161/278 (57%), Gaps = 24/278 (8%)
Query 2 RSLPHFSKFDYSPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRL 61
R+LPHF K DY PESRGFVE+SYL GL P E +FHAMGGREG+IDTA KT+ETGYIQRRL
Sbjct 797 RTLPHFIKDDYGPESRGFVENSYLAGLTPSEFYFHAMGGREGLIDTAVKTAETGYIQRRL 856
Query 62 MKAMEDVKVSYDRTVRNSAQEVLQFLYGEDGMSGEYVEDQVIELMQMDAEKIRRVYRHDL 121
+KAME V V+YD TVRNS +++Q YGEDG+ GE VE Q + +++ + + ++ D
Sbjct 857 IKAMESVMVNYDGTVRNSVGQLIQLRYGEDGLCGELVEFQNMPTVKLSNKSFEKRFKFD- 915
Query 122 DQESYGKGWILNEELRTKLFVD---------FSAQQVLEDEYEAIKEDKLRLCKSIFPDG 172
W NE L K+F D A Q LE E++ + D+ L + IFP+G
Sbjct 916 --------W-SNERLMKKVFTDDVIKEMTDSSEAIQELEAEWDRLVSDRDSL-RQIFPNG 965
Query 173 EVKQHIPINIMRLLEFAKAQFPPSVEQLKKQDPVAIAKSVSELLNDKLIVVKQTCKADTI 232
E K +P N+ R++ + F + P+ + K V LL +IV D I
Sbjct 966 ESKVVLPCNLQRMIWNVQKIFHINKRLPTDLSPIRVIKGVKTLLERCVIVTGN----DRI 1021
Query 233 SAEVQENATVFMKAHLRTVLNSRRLMEREQLGVKAVHW 270
S + ENAT+ + +R+ L ++ + E +L +A W
Sbjct 1022 SKQANENATLLFQCLIRSTLCTKYVSEEFRLSTEAFEW 1059
> YDL140c
Length=1733
Score = 202 bits (514), Expect = 7e-52, Method: Compositional matrix adjust.
Identities = 113/272 (41%), Positives = 165/272 (60%), Gaps = 10/272 (3%)
Query 1 DRSLPHFSKFDYSPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRR 60
DR+LPHFSK DYSPES+GFVE+SYL GL PQE FFHAMGGREG+IDTA KT+ETGYIQRR
Sbjct 781 DRTLPHFSKDDYSPESKGFVENSYLRGLTPQEFFFHAMGGREGLIDTAVKTAETGYIQRR 840
Query 61 LMKAMEDVKVSYDRTVRNSAQEVLQFLYGEDGMSGEYVEDQVIELMQMDAEKIRRVYRHD 120
L+KA+ED+ V YD T RNS V+QF+YGEDGM ++E Q ++ + + YR D
Sbjct 841 LVKALEDIMVHYDNTTRNSLGNVIQFIYGEDGMDAAHIEKQSLDTIGGSDAAFEKRYRVD 900
Query 121 LDQESYGKGWILNEELRTKLFVDFSAQQVLEDEYEAIKEDKLRLCKSIFPDGEVKQHIPI 180
L + L E +++ D Q +L++EY+ + +D+ + + +F DGE +P+
Sbjct 901 LLNTDHTLDPSLLES-GSEILGDLKLQVLLDEEYKQLVKDR-KFLREVFVDGEANWPLPV 958
Query 181 NIMRLLEFAKAQFPPSVEQLKKQDPVA--IAKSVSELLNDKLIVVKQTCKADTISAEVQE 238
NI R+++ A+ F ++ K D I V + L + L+V++ + I Q
Sbjct 959 NIRRIIQNAQQTF--HIDHTKPSDLTIKDIVLGVKD-LQENLLVLR---GKNEIIQNAQR 1012
Query 239 NATVFMKAHLRTVLNSRRLMEREQLGVKAVHW 270
+A LR+ L +RR+++ +L +A W
Sbjct 1013 DAVTLFCCLLRSRLATRRVLQEYRLTKQAFDW 1044
> ECU03g0290
Length=1599
Score = 144 bits (362), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 78/156 (50%), Positives = 103/156 (66%), Gaps = 7/156 (4%)
Query 2 RSLPHFSKFDYSPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRL 61
R+LPHF K DY+ +SRGFVE+SYLTGL P+E FFHAMGGREG+IDTA KT+ETGYIQRRL
Sbjct 776 RTLPHFVKDDYTGKSRGFVENSYLTGLDPEEFFFHAMGGREGLIDTAIKTAETGYIQRRL 835
Query 62 MKAMEDVKVSYDRTVRNSAQEVLQFLYGEDGMSGEYVEDQVIELMQMDAEKIRRVYRHDL 121
+KA+ED V D +VR+ V Q YGEDG ++E Q ++D + + Y D+
Sbjct 836 VKALEDAIVRQDESVRSGNGLVYQIKYGEDGFDATFLESQ-----KVDVKNFTKRYYIDM 890
Query 122 --DQESYGKGWILNEELRTKLFVDFSAQQVLEDEYE 155
+E K ++EE+ L D Q++L+ EYE
Sbjct 891 FGTEELEIKHGQVSEEVYGMLSSDVDLQKLLDQEYE 926
> At5g60040
Length=1328
Score = 132 bits (333), Expect = 5e-31, Method: Compositional matrix adjust.
Identities = 69/169 (40%), Positives = 100/169 (59%), Gaps = 15/169 (8%)
Query 1 DRSLPHFSKFDYSPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRR 60
DRSLPHF + SP ++GFV +S+ +GL E FFH MGGREG++DTA KT+ TGY+ RR
Sbjct 792 DRSLPHFPRMSKSPAAKGFVANSFYSGLTATEFFFHTMGGREGLVDTAVKTASTGYMSRR 851
Query 61 LMKAMEDVKVSYDRTVRNSAQEVLQFLYGEDGMSGEYVEDQVIELMQMDAEKIRRVYRHD 120
LMKA+ED+ V YD TVRN++ +LQF YG+DGM +E + + ++ ++ +
Sbjct 852 LMKALEDLLVHYDNTVRNASGCILQFTYGDDGMDPALMEGK--DGAPLNFNRLFLKVQAT 909
Query 121 LDQESYGKGWILNEELRTKLFVDFSAQQVLEDEYEAIKEDKLRLCKSIF 169
S+ ++ +EEL K E E ++ DK R+C F
Sbjct 910 CPPRSH-HTYLSSEELSQKF------------EEELVRHDKSRVCTDAF 945
> CE04196
Length=1388
Score = 131 bits (329), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 55/99 (55%), Positives = 78/99 (78%), Gaps = 0/99 (0%)
Query 1 DRSLPHFSKFDYSPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRR 60
+RSLPHF + +PE++GFV +S+ +GL P E FFH MGGREG++DTA KT+ETGY+QRR
Sbjct 811 ERSLPHFERKKKTPEAKGFVANSFYSGLTPTEFFFHTMGGREGLVDTAVKTAETGYMQRR 870
Query 61 LMKAMEDVKVSYDRTVRNSAQEVLQFLYGEDGMSGEYVE 99
L+K +ED+ SYD TVR+S +V++F++GEDG+ +E
Sbjct 871 LVKCLEDLCASYDGTVRSSVGDVIEFVFGEDGLDPAMME 909
> SPBC651.08c
Length=1405
Score = 130 bits (328), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 61/123 (49%), Positives = 85/123 (69%), Gaps = 3/123 (2%)
Query 1 DRSLPHFSKFDYSPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRR 60
DRSLPHF K P ++GFV +S+ +GL P E FHA+ GREG++DTA KT+ETGY+ RR
Sbjct 809 DRSLPHFHKNSKHPLAKGFVSNSFYSGLTPTEFLFHAISGREGLVDTAVKTAETGYMSRR 868
Query 61 LMKAMEDVKVSYDRTVRNSAQEVLQFLYGEDGMSGEYVE--DQVIELMQMDAEKIRRVY- 117
LMK++ED+ +YD TVR+S +V+QF+YG+DG+ Y+E Q +E + + Y
Sbjct 869 LMKSLEDLSSAYDGTVRSSNSDVVQFVYGDDGLDPTYMEGDGQAVEFKRTWIHSVNLNYD 928
Query 118 RHD 120
RHD
Sbjct 929 RHD 931
> YOR116c
Length=1460
Score = 129 bits (323), Expect = 8e-30, Method: Compositional matrix adjust.
Identities = 54/93 (58%), Positives = 71/93 (76%), Gaps = 0/93 (0%)
Query 1 DRSLPHFSKFDYSPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRR 60
DRSLPHF K +P+S+GFV +S+ +GL P E FHA+ GREG++DTA KT+ETGY+ RR
Sbjct 829 DRSLPHFPKNSKTPQSKGFVRNSFFSGLSPPEFLFHAISGREGLVDTAVKTAETGYMSRR 888
Query 61 LMKAMEDVKVSYDRTVRNSAQEVLQFLYGEDGM 93
LMK++ED+ YD TVR SA ++QF YG DG+
Sbjct 889 LMKSLEDLSCQYDNTVRTSANGIVQFTYGGDGL 921
> ECU07g0090
Length=1349
Score = 127 bits (319), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 60/109 (55%), Positives = 79/109 (72%), Gaps = 1/109 (0%)
Query 1 DRSLPHFSKFDYSPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRR 60
+R LPHF + +PES+GFV +S+ GL E FFHA+ GREG++DTA KT+ETGY+QRR
Sbjct 813 ERCLPHFKRGSRTPESKGFVLNSFFDGLTSPEFFFHAVSGREGLVDTAVKTAETGYMQRR 872
Query 61 LMKAMEDVKVSYDRTVRNSAQEVLQFLYGEDGMSGEYVE-DQVIELMQM 108
LMKA+ED+ + YD +VRNS EV+QF YGED + E D+ I L Q+
Sbjct 873 LMKALEDLSIQYDGSVRNSNMEVVQFAYGEDQIDPAMSEGDESINLEQV 921
> Hs5902062
Length=1391
Score = 125 bits (313), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 53/101 (52%), Positives = 75/101 (74%), Gaps = 0/101 (0%)
Query 1 DRSLPHFSKFDYSPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRR 60
+RSLPHF K P ++GFV +S+ +GL P E FFH M GREG++DTA KT+ETGY+QRR
Sbjct 815 NRSLPHFEKHSKLPAAKGFVANSFYSGLTPTEFFFHTMAGREGLVDTAVKTAETGYMQRR 874
Query 61 LMKAMEDVKVSYDRTVRNSAQEVLQFLYGEDGMSGEYVEDQ 101
L+K++ED+ YD TVR+S +++QF+YG DG+ +E +
Sbjct 875 LVKSLEDLCSQYDLTVRSSTGDIIQFIYGGDGLDPAAMEGK 915
> 7293125
Length=1259
Score = 124 bits (311), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 51/93 (54%), Positives = 71/93 (76%), Gaps = 0/93 (0%)
Query 1 DRSLPHFSKFDYSPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRR 60
+R+LPHF + P +RGFV++S+ +GL P E FFH M GREG++DTA KT+ETGY+QRR
Sbjct 689 NRALPHFERHSAIPAARGFVQNSFYSGLTPTEFFFHTMAGREGLVDTAVKTAETGYLQRR 748
Query 61 LMKAMEDVKVSYDRTVRNSAQEVLQFLYGEDGM 93
L+K +ED+ V YD TVRN+ E++ +YG DG+
Sbjct 749 LVKCLEDLVVHYDGTVRNAVNEMVDTIYGGDGL 781
> 7303170
Length=1642
Score = 121 bits (304), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 52/92 (56%), Positives = 71/92 (77%), Gaps = 0/92 (0%)
Query 2 RSLPHFSKFDYSPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRL 61
+SLP F+ F+ SP+S GF++ ++TG+ PQ+ FFH M GREG+IDTA KTS +GY+QR L
Sbjct 911 KSLPSFTSFETSPKSGGFIDGRFMTGIQPQDFFFHCMAGREGLIDTAVKTSRSGYLQRCL 970
Query 62 MKAMEDVKVSYDRTVRNSAQEVLQFLYGEDGM 93
+K +E + V YD TVR+S V+QFLYGEDG+
Sbjct 971 IKHLEGLSVHYDLTVRDSDNSVVQFLYGEDGL 1002
> CE22157
Length=1737
Score = 120 bits (300), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 52/92 (56%), Positives = 69/92 (75%), Gaps = 0/92 (0%)
Query 2 RSLPHFSKFDYSPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRL 61
R+LP F FD SP + G+++ +LTG+ PQELFFH M GREG+IDTA KTS +GY+QR +
Sbjct 969 RTLPSFRCFDPSPRAGGYIDQRFLTGMNPQELFFHTMAGREGLIDTAVKTSRSGYLQRCI 1028
Query 62 MKAMEDVKVSYDRTVRNSAQEVLQFLYGEDGM 93
+K +E ++V YD TVR+ V+QF YGEDGM
Sbjct 1029 IKHLEGIRVHYDSTVRDHDGSVIQFRYGEDGM 1060
> At3g57660
Length=1670
Score = 117 bits (293), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 49/92 (53%), Positives = 70/92 (76%), Gaps = 0/92 (0%)
Query 2 RSLPHFSKFDYSPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRL 61
++LP F +D+SP + GF+ +L+GL PQE +FH M GREG++DTA KTS +GY+QR L
Sbjct 977 KTLPCFHPWDWSPRAGGFISDRFLSGLRPQEYYFHCMAGREGLVDTAVKTSRSGYLQRCL 1036
Query 62 MKAMEDVKVSYDRTVRNSAQEVLQFLYGEDGM 93
MK +E +KV+YD TVR++ ++QF YGEDG+
Sbjct 1037 MKNLESLKVNYDCTVRDADGSIIQFQYGEDGV 1068
> Hs7661686
Length=1717
Score = 114 bits (285), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 49/92 (53%), Positives = 68/92 (73%), Gaps = 0/92 (0%)
Query 2 RSLPHFSKFDYSPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRL 61
+SLP F ++++P + GFV +LTG+ P E FFH M GREG++DTA KTS +GY+QR +
Sbjct 933 KSLPCFEPYEFTPRAGGFVTGRFLTGIKPPEFFFHCMAGREGLVDTAVKTSRSGYLQRCI 992
Query 62 MKAMEDVKVSYDRTVRNSAQEVLQFLYGEDGM 93
+K +E + V YD TVR+S V+QFLYGEDG+
Sbjct 993 IKHLEGLVVQYDLTVRDSDGSVVQFLYGEDGL 1024
> SPBC4C3.05c
Length=1689
Score = 110 bits (275), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 49/92 (53%), Positives = 66/92 (71%), Gaps = 0/92 (0%)
Query 2 RSLPHFSKFDYSPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRL 61
+SLP F ++ S +S GF+ +LTG+ PQE +FH M GREG+IDTA KTS +GY+QR L
Sbjct 977 KSLPSFVPYETSAKSGGFIASRFLTGIAPQEYYFHCMAGREGLIDTAVKTSRSGYLQRCL 1036
Query 62 MKAMEDVKVSYDRTVRNSAQEVLQFLYGEDGM 93
MK +E + V YD TVR+S ++QF YGED +
Sbjct 1037 MKHLEGLCVQYDHTVRDSDGSIVQFHYGEDSL 1068
> ECU04g1400
Length=1395
Score = 107 bits (268), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 53/103 (51%), Positives = 73/103 (70%), Gaps = 6/103 (5%)
Query 2 RSLPHFSKFDYSPESRGFVEHSYLTGL-LPQELFFHAMGGREGIIDTACKTSETGYIQRR 60
++LP F+ D SP S G++ H +LTG+ LPQ FFH M GREG+IDTA KT+ +GY+QR
Sbjct 802 KTLPCFAALDPSPSSGGYIYHRFLTGIDLPQ-YFFHCMAGREGLIDTAIKTANSGYLQRC 860
Query 61 LMKAMEDVKVSYDRTVRNSAQEVLQFLYGEDGMS---GEYVED 100
L+K ME+ KV YD +VR + V+QF+YGEDG+ Y++D
Sbjct 861 LIKHMEEAKVEYDMSVR-IGKRVIQFMYGEDGLDCTKSSYLDD 902
> YOR341w
Length=1664
Score = 99.0 bits (245), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 41/92 (44%), Positives = 64/92 (69%), Gaps = 0/92 (0%)
Query 2 RSLPHFSKFDYSPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRL 61
++LP F ++ + G+V+ + +G+ PQE +FH M GREG+IDTA KTS +GY+QR L
Sbjct 964 KTLPSFKPYETDAMAGGYVKGRFYSGIKPQEYYFHCMAGREGLIDTAVKTSRSGYLQRCL 1023
Query 62 MKAMEDVKVSYDRTVRNSAQEVLQFLYGEDGM 93
K +E V VSYD ++R++ ++QF+YG D +
Sbjct 1024 TKQLEGVHVSYDNSIRDADGTLVQFMYGGDAI 1055
> At1g63020_1
Length=1316
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/76 (40%), Positives = 44/76 (57%), Gaps = 3/76 (3%)
Query 18 GFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRLMKAMEDVKVSYDRTVR 77
G +E+S+LTGL P E F H++ R+ + G + RRLM M D+ +YD TVR
Sbjct 794 GVIENSFLTGLNPLESFVHSVTSRDSSF--SGNADLPGTLSRRLMFFMRDIYAAYDGTVR 851
Query 78 NS-AQEVLQFLYGEDG 92
NS +++QF Y DG
Sbjct 852 NSFGNQLVQFTYETDG 867
> AtCh013
Length=1376
Score = 42.0 bits (97), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 28/44 (63%), Gaps = 0/44 (0%)
Query 27 GLLPQELFFHAMGGREGIIDTACKTSETGYIQRRLMKAMEDVKV 70
GL E G R+G++DTA +TS+ GY+ RRL++ ++ + V
Sbjct 172 GLSLTEYIISCYGARKGVVDTAVRTSDAGYLTRRLVEVVQHIVV 215
> 7290249
Length=1398
Score = 31.2 bits (69), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 23/94 (24%), Positives = 43/94 (45%), Gaps = 5/94 (5%)
Query 28 LLPQELFFHAMGGREGIIDTACKTSETGYIQRRLMKAMEDVKVSY---DRTVRNSAQEVL 84
LL Q+LF G G T+ G + L+K E ++V + +RT+ V+
Sbjct 96 LLAQKLFASGGGSTPGPSPTSSAVGAGGISGKDLLKLKEPMRVGFYDIERTIGKGNFAVV 155
Query 85 QFLYGEDGMSGEYVEDQVIELMQMDAEKIRRVYR 118
+ ++ V ++I+ Q+D +++VYR
Sbjct 156 KL--ARHRITKNEVAIKIIDKSQLDQTNLQKVYR 187
> CE25973
Length=334
Score = 29.6 bits (65), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 25/49 (51%), Gaps = 2/49 (4%)
Query 76 VRNSAQEVLQFLYGEDGMSGEYVEDQVIELMQMDAEKIRRVYRHDLDQE 124
V NS + FL DG + +Y+ ++IE ++D E I R+ D E
Sbjct 146 VVNSLS--ILFLTSPDGFTDDYLRAEIIEKYELDIENIARLVMIPFDTE 192
Lambda K H
0.319 0.135 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5773141498
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40