bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
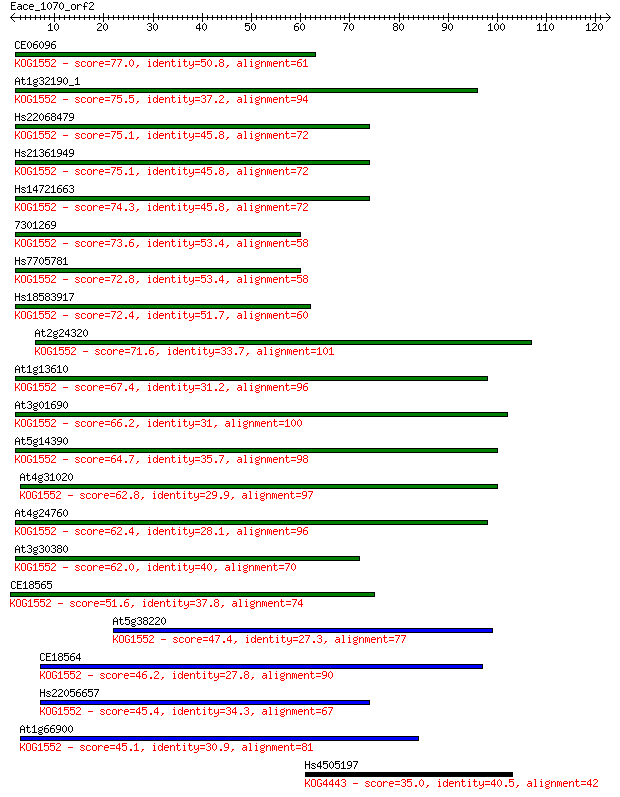
Query= Eace_1070_orf2
Length=122
Score E
Sequences producing significant alignments: (Bits) Value
CE06096 77.0 8e-15
At1g32190_1 75.5 2e-14
Hs22068479 75.1 3e-14
Hs21361949 75.1 3e-14
Hs14721663 74.3 5e-14
7301269 73.6 9e-14
Hs7705781 72.8 2e-13
Hs18583917 72.4 2e-13
At2g24320 71.6 4e-13
At1g13610 67.4 7e-12
At3g01690 66.2 1e-11
At5g14390 64.7 4e-11
At4g31020 62.8 1e-10
At4g24760 62.4 2e-10
At3g30380 62.0 2e-10
CE18565 51.6 4e-07
At5g38220 47.4 7e-06
CE18564 46.2 1e-05
Hs22056657 45.4 3e-05
At1g66900 45.1 3e-05
Hs4505197 35.0 0.030
> CE06096
Length=405
Score = 77.0 bits (188), Expect = 8e-15, Method: Composition-based stats.
Identities = 31/61 (50%), Positives = 42/61 (68%), Gaps = 0/61 (0%)
Query 2 DLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVA 61
D F +I+K+ +VKCP IHG +DEV+ HG+ +Y+R V PLWV GAGHN+VE+ A
Sbjct 306 DAFPSIEKVPRVKCPTLVIHGTDDEVIDFSHGVSIYERCPTSVEPLWVPGAGHNDVELHA 365
Query 62 G 62
Sbjct 366 A 366
> At1g32190_1
Length=289
Score = 75.5 bits (184), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 35/94 (37%), Positives = 57/94 (60%), Gaps = 2/94 (2%)
Query 2 DLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVA 61
D++ N+ KIKKVKCPV IHG D+VV HG L+K A+ PLW+ G GH N+EI
Sbjct 198 DIYSNVNKIKKVKCPVLVIHGTEDDVVNWLHGNRLWKMAKEPYEPLWIKGGGHCNLEIYP 257
Query 62 GREFLLAISRFLRLLQQQQQQQQQQQQQQEQQQK 95
+++ + RF++ ++ + + + QE +++
Sbjct 258 --DYIRHLYRFIQDMENTTTKSRLKTIWQEIRRR 289
> Hs22068479
Length=161
Score = 75.1 bits (183), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 33/72 (45%), Positives = 47/72 (65%), Gaps = 2/72 (2%)
Query 2 DLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVA 61
D F NI+K+ K+ PV IHG DEV+ HG+ LY+R V PLWV GAGHN++E+ +
Sbjct 83 DAFPNIEKVSKITSPVLIIHGMEDEVIDFSHGLALYERCPKAVEPLWVEGAGHNDIELYS 142
Query 62 GREFLLAISRFL 73
++L + RF+
Sbjct 143 --QYLERLRRFI 152
> Hs21361949
Length=361
Score = 75.1 bits (183), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 33/72 (45%), Positives = 47/72 (65%), Gaps = 2/72 (2%)
Query 2 DLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVA 61
D F NI+K+ K+ PV IHG DEV+ HG+ LY+R V PLWV GAGHN++E+ +
Sbjct 283 DAFPNIEKVSKITSPVLIIHGTEDEVIDFSHGLALYERCPKAVEPLWVEGAGHNDIELYS 342
Query 62 GREFLLAISRFL 73
++L + RF+
Sbjct 343 --QYLERLRRFI 352
> Hs14721663
Length=310
Score = 74.3 bits (181), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 33/72 (45%), Positives = 47/72 (65%), Gaps = 2/72 (2%)
Query 2 DLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVA 61
D F NI+K+ K+ PV IHG DEV+ HG+ LY+R V PLWV GAGHN++E+ +
Sbjct 232 DAFPNIEKVSKITSPVLIIHGREDEVIDFSHGLALYERCPKAVEPLWVEGAGHNDIELYS 291
Query 62 GREFLLAISRFL 73
++L + RF+
Sbjct 292 --QYLERLRRFI 301
> 7301269
Length=286
Score = 73.6 bits (179), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 31/58 (53%), Positives = 39/58 (67%), Gaps = 0/58 (0%)
Query 2 DLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEI 59
D F +I K+ KVK PV IHG +DEV+ HGI +Y+R V P WV GAGHN+VE+
Sbjct 211 DAFPSIDKVAKVKAPVLVIHGTDDEVIDFSHGIGIYERCPKTVEPFWVEGAGHNDVEL 268
> Hs7705781
Length=293
Score = 72.8 bits (177), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 31/58 (53%), Positives = 39/58 (67%), Gaps = 0/58 (0%)
Query 2 DLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEI 59
D F NI KI K+ PV IHG DEV+ HG+ L++R + V PLWV GAGHN+VE+
Sbjct 212 DAFPNIDKISKITSPVLIIHGTEDEVIDFSHGLALFERCQRPVEPLWVEGAGHNDVEL 269
> Hs18583917
Length=329
Score = 72.4 bits (176), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 31/60 (51%), Positives = 39/60 (65%), Gaps = 0/60 (0%)
Query 2 DLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVA 61
D F +I KI KV PV IHG DEV+ HG+ +Y+R V PLWV GAGHN++E+ A
Sbjct 253 DAFPSIDKISKVTSPVLVIHGTEDEVIDFSHGLAMYERCPRAVEPLWVEGAGHNDIELYA 312
> At2g24320
Length=316
Score = 71.6 bits (174), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 34/104 (32%), Positives = 57/104 (54%), Gaps = 5/104 (4%)
Query 6 NIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVAGREF 65
NI KI+ V CPV IHG D++V + HG L++ A+ + PLWV G GH N+E E+
Sbjct 215 NIDKIRHVTCPVLVIHGTKDDIVNMSHGKRLWELAKDKYDPLWVKGGGHCNLETYP--EY 272
Query 66 LLAISRFLRLLQQ---QQQQQQQQQQQQEQQQKQQRRLLGAAAR 106
+ + +F+ +++ +QQ + ++ +Q R L + R
Sbjct 273 IKHMRKFMNAMEKLALNNPPNKQQNDEPSIKETKQNRCLRFSKR 316
> At1g13610
Length=351
Score = 67.4 bits (163), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 30/96 (31%), Positives = 55/96 (57%), Gaps = 2/96 (2%)
Query 2 DLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVA 61
D++ NI KI V+CPV IHG +D+VV I HG L+ + + PLW+ G GH+++E+
Sbjct 185 DIYKNIDKIHLVECPVLVIHGTDDDVVNISHGKHLWGLCKEKYEPLWLKGRGHSDIEMSP 244
Query 62 GREFLLAISRFLRLLQQQQQQQQQQQQQQEQQQKQQ 97
E+L + +F+ +++ + ++Q + +
Sbjct 245 --EYLPHLRKFISAIEKLPVPKFRRQSLANDHKNDK 278
> At3g01690
Length=361
Score = 66.2 bits (160), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 31/100 (31%), Positives = 53/100 (53%), Gaps = 2/100 (2%)
Query 2 DLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVA 61
D++ NI KI V CPV IHG +DEVV HG +L++ + + PLWV G H ++E
Sbjct 189 DIYKNIDKIPYVDCPVLIIHGTSDEVVDCSHGKQLWELCKDKYEPLWVKGGNHCDLEHYP 248
Query 62 GREFLLAISRFLRLLQQQQQQQQQQQQQQEQQQKQQRRLL 101
E++ + +F+ +++ + Q + + RR +
Sbjct 249 --EYIRHLKKFIATVERLPCPRMSSDQSERVRDAPPRRSM 286
> At5g14390
Length=369
Score = 64.7 bits (156), Expect = 4e-11, Method: Composition-based stats.
Identities = 35/103 (33%), Positives = 53/103 (51%), Gaps = 5/103 (4%)
Query 2 DLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVE--- 58
D+F NI KI V CPV IHG DEVV HG +L++ ++ + PLW+ G H ++E
Sbjct 189 DIFKNIDKIPLVNCPVLVIHGTCDEVVDCSHGKQLWELSKEKYEPLWLEGGNHCDLEHYP 248
Query 59 --IVAGREFLLAISRFLRLLQQQQQQQQQQQQQQEQQQKQQRR 99
I ++F+ + R L Q ++Q + +Q RR
Sbjct 249 EYIKHLKKFITTVERDLSSRASTAQLEKQSSDLEMPRQSVDRR 291
> At4g31020
Length=307
Score = 62.8 bits (151), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 29/97 (29%), Positives = 51/97 (52%), Gaps = 2/97 (2%)
Query 3 LFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVAG 62
++ NI KI+ V V IHG NDE+V + HG L++ A+ + PLWV G GH N+E
Sbjct 203 IYSNIDKIRHVNSQVLVIHGTNDEIVDLSHGKRLWELAKEKYDPLWVKGGGHCNLETYP- 261
Query 63 REFLLAISRFLRLLQQQQQQQQQQQQQQEQQQKQQRR 99
E++ + +F+ +++ +Q + + +
Sbjct 262 -EYIKHLKKFVNAMEKLSLTNPPPKQLTNEPSITETK 297
> At4g24760
Length=365
Score = 62.4 bits (150), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 27/96 (28%), Positives = 54/96 (56%), Gaps = 2/96 (2%)
Query 2 DLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVA 61
D++ NI KI V+CPV IHG D+VV HG +L++ + + PLW+ G H ++E+
Sbjct 189 DIYKNIDKITLVRCPVLVIHGTADDVVDFSHGKQLWELCQEKYEPLWLKGGNHCDLELFP 248
Query 62 GREFLLAISRFLRLLQQQQQQQQQQQQQQEQQQKQQ 97
E++ + +F+ +++ ++ ++ + +Q
Sbjct 249 --EYIGHLKKFVSAVEKSASKRNSSFSRRSMEGCEQ 282
> At3g30380
Length=399
Score = 62.0 bits (149), Expect = 2e-10, Method: Composition-based stats.
Identities = 28/75 (37%), Positives = 46/75 (61%), Gaps = 5/75 (6%)
Query 2 DLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVA 61
D++ N++KI VKCPV IHG +D+VV HG +L++ + + PLW+ G H ++E+
Sbjct 188 DIYKNVEKISFVKCPVLVIHGTSDDVVNWSHGKQLFELCKEKYEPLWIKGGNHCDLELYP 247
Query 62 G-----REFLLAISR 71
R+F+ AI +
Sbjct 248 QYIKHLRKFVSAIEK 262
> CE18565
Length=305
Score = 51.6 bits (122), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 28/76 (36%), Positives = 41/76 (53%), Gaps = 4/76 (5%)
Query 1 GDLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIV 60
D F + KI + V HG DEV+P+ HG+ LY++ + V PL V GA H+ I+
Sbjct 203 ADSFKSFDKINNIDTRVLICHGDVDEVIPLSHGLALYEKLKNPVPPLIVHGANHHT--IL 260
Query 61 AGR--EFLLAISRFLR 74
+G+ I+ FLR
Sbjct 261 SGKYIHVFTRIANFLR 276
> At5g38220
Length=320
Score = 47.4 bits (111), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 41/77 (53%), Gaps = 2/77 (2%)
Query 22 GANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVAGREFLLAISRFLRLLQQQQQ 81
G DEVV HG +L++ ++ + PLWV G GH N+E+ EF+ + +++ + + +
Sbjct 191 GTADEVVDCSHGKQLWELSKEKYEPLWVSGGGHCNLELYP--EFIKHLKKYVISISKGPR 248
Query 82 QQQQQQQQQEQQQKQQR 98
+ + +KQ +
Sbjct 249 TGSNKTATTDAAKKQSK 265
> CE18564
Length=335
Score = 46.2 bits (108), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/90 (27%), Positives = 46/90 (51%), Gaps = 3/90 (3%)
Query 7 IKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVAGREFL 66
I KI + V HG +D+ +P+ HG+ LY+ + V PL V GA H++ I++G E++
Sbjct 245 IDKICHINTRVLICHGDHDQRIPMTHGMALYENLKNPVPPLIVHGANHHS--IISG-EYI 301
Query 67 LAISRFLRLLQQQQQQQQQQQQQQEQQQKQ 96
+R ++ + + Q + K+
Sbjct 302 EVFTRIASFMRNETLLSCRANQIESSSSKK 331
> Hs22056657
Length=124
Score = 45.4 bits (106), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 36/67 (53%), Gaps = 13/67 (19%)
Query 7 IKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVAGREFL 66
I+K+ K+ PV IHG DE +R V PLWV GAGH ++++ + ++L
Sbjct 62 IQKVSKITSPVLIIHGTKDE-----------ERCPKAVEPLWVEGAGHKDIQLYS--QYL 108
Query 67 LAISRFL 73
+ RF+
Sbjct 109 ERLRRFI 115
> At1g66900
Length=256
Score = 45.1 bits (105), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 46/83 (55%), Gaps = 4/83 (4%)
Query 3 LFVNIKKIKKVKCPV-FCIH-GANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIV 60
+ ++ + VKC F I+ G DEVV HG L++ ++ + PLW+ G GH ++E+
Sbjct 173 ILSGMRVLYPVKCTYWFDIYKGTADEVVDWSHGKRLWELSKEKYEPLWISGGGHCDLELY 232
Query 61 AGREFLLAISRFLRLLQQQQQQQ 83
+F+ + +F+ L +Q +Q
Sbjct 233 P--DFIRHLKKFVVSLGNKQAEQ 253
> Hs4505197
Length=5262
Score = 35.0 bits (79), Expect = 0.030, Method: Composition-based stats.
Identities = 17/42 (40%), Positives = 26/42 (61%), Gaps = 0/42 (0%)
Query 61 AGREFLLAISRFLRLLQQQQQQQQQQQQQQEQQQKQQRRLLG 102
AGREF A + L+L+ +QQ + Q+Q Q +QQK+ L+
Sbjct 3277 AGREFPEADAEKLKLVTEQQSKIQKQLDQVRKQQKEHTNLMA 3318
Lambda K H
0.322 0.135 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194805952
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40