bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1057_orf1
Length=188
Score E
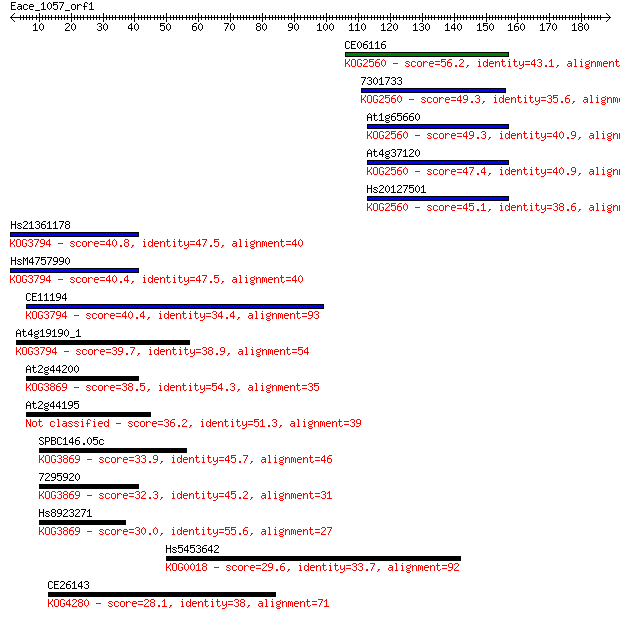
Sequences producing significant alignments: (Bits) Value
CE06116 56.2 4e-08
7301733 49.3 4e-06
At1g65660 49.3 5e-06
At4g37120 47.4 2e-05
Hs20127501 45.1 8e-05
Hs21361178 40.8 0.002
HsM4757990 40.4 0.002
CE11194 40.4 0.002
At4g19190_1 39.7 0.003
At2g44200 38.5 0.008
At2g44195 36.2 0.036
SPBC146.05c 33.9 0.18
7295920 32.3 0.52
Hs8923271 30.0 2.7
Hs5453642 29.6 4.2
CE26143 28.1 9.5
> CE06116
Length=647
Score = 56.2 bits (134), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 22/51 (43%), Positives = 33/51 (64%), Gaps = 2/51 (3%)
Query 106 KLSKSTLYDEDAFTQGHTSVFGSLYSKEENKWGYRCCGSLDRSAPCTANQG 156
K++ S+ + ED + Q HTSVFGS + E +WGY+CC +++ CT QG
Sbjct 388 KVAISSRFKEDIYPQNHTSVFGSFW--REGRWGYKCCHQFVKNSYCTGKQG 436
> 7301733
Length=527
Score = 49.3 bits (116), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 29/45 (64%), Gaps = 2/45 (4%)
Query 111 TLYDEDAFTQGHTSVFGSLYSKEENKWGYRCCGSLDRSAPCTANQ 155
++Y+ED + HT+V+GS ++ +WGY+CC S +++ C Q
Sbjct 388 SIYEEDVYINNHTTVWGSFWNA--GRWGYKCCKSFIKNSYCVGMQ 430
> At1g65660
Length=535
Score = 49.3 bits (116), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 30/44 (68%), Gaps = 2/44 (4%)
Query 113 YDEDAFTQGHTSVFGSLYSKEENKWGYRCCGSLDRSAPCTANQG 156
Y+ED HTSV+GS + ++++WGY+CC + R++ CT + G
Sbjct 390 YEEDVHANNHTSVWGSYW--KDHQWGYKCCQQIIRNSYCTGSAG 431
> At4g37120
Length=538
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 29/44 (65%), Gaps = 2/44 (4%)
Query 113 YDEDAFTQGHTSVFGSLYSKEENKWGYRCCGSLDRSAPCTANQG 156
Y+ED HTSV+GS + ++++WGY+CC R++ CT + G
Sbjct 392 YEEDVHANNHTSVWGSWW--KDHQWGYKCCQQTIRNSYCTGSAG 433
> Hs20127501
Length=586
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 26/44 (59%), Gaps = 2/44 (4%)
Query 113 YDEDAFTQGHTSVFGSLYSKEENKWGYRCCGSLDRSAPCTANQG 156
Y+ED HT ++GS + +E +WGY+CC S + + CT G
Sbjct 419 YEEDVKIHNHTHIWGSYW--KEGRWGYKCCHSFFKYSYCTGEAG 460
> Hs21361178
Length=451
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/40 (47%), Positives = 28/40 (70%), Gaps = 0/40 (0%)
Query 1 KKSAAFINHKPFHPGNYQNQEKVWLAEQKLKEEQKKEKEL 40
K A F+ K FHP + N +KVW+AEQK+ ++KK++EL
Sbjct 3 KSFANFMCKKDFHPASKSNIKKVWMAEQKISYDKKKQEEL 42
> HsM4757990
Length=450
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/40 (47%), Positives = 28/40 (70%), Gaps = 0/40 (0%)
Query 1 KKSAAFINHKPFHPGNYQNQEKVWLAEQKLKEEQKKEKEL 40
K A F+ K FHP + N +KVW+AEQK+ ++KK++EL
Sbjct 3 KSFANFMCKKDFHPASKSNIKKVWMAEQKISYDKKKQEEL 42
> CE11194
Length=560
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 32/100 (32%), Positives = 53/100 (53%), Gaps = 17/100 (17%)
Query 6 FINHKPFHPGNYQNQEKVWLAEQKLKEEQKKEKELEERRREEVKI-----EEL--RRALR 58
F++ K FHP ++N + VW A QK K LE++R+EE+++ +E+ +AL
Sbjct 8 FMSKKDFHPSAFRNLKMVWEARQK--------KSLEDKRQEELRVAYEKEQEILNNKALL 59
Query 59 GEAKAALQLQKKEEAVSPAEELRRKAKEAARLQALQRKHQ 98
G+ KA + L +A PA +R+ + QRK+Q
Sbjct 60 GDEKAKMGLSFMYDA--PAGMTKREEPKEEPKFEWQRKYQ 97
> At4g19190_1
Length=658
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 30/54 (55%), Gaps = 8/54 (14%)
Query 3 SAAFINHKPFHPGNYQNQEKVWLAEQKLKEEQKKEKELEERRREEVKIEELRRA 56
S +F+N KP+HP +Y NQ + W+AEQ + +RR EEV E + R
Sbjct 39 SHSFLNQKPWHPLSYPNQRRKWIAEQTHAQ--------HDRRAEEVAREVIHRT 84
> At2g44200
Length=493
Score = 38.5 bits (88), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 19/35 (54%), Positives = 25/35 (71%), Gaps = 0/35 (0%)
Query 6 FINHKPFHPGNYQNQEKVWLAEQKLKEEQKKEKEL 40
F+N K +H G+ +N E VW AEQK + EQKK +EL
Sbjct 5 FLNKKGWHTGSLRNIETVWKAEQKQEAEQKKLEEL 39
> At2g44195
Length=107
Score = 36.2 bits (82), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 20/39 (51%), Positives = 27/39 (69%), Gaps = 0/39 (0%)
Query 6 FINHKPFHPGNYQNQEKVWLAEQKLKEEQKKEKELEERR 44
F+N K +H G+ +N EKVW AEQK + EQ+K +E E R
Sbjct 5 FLNKKGWHTGSIRNVEKVWKAEQKHEAEQEKIEERSEFR 43
> SPBC146.05c
Length=376
Score = 33.9 bits (76), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 21/46 (45%), Positives = 28/46 (60%), Gaps = 0/46 (0%)
Query 10 KPFHPGNYQNQEKVWLAEQKLKEEQKKEKELEERRREEVKIEELRR 55
K +HP +NQEKVW EQ KEE K+ ++L EE ++ EL R
Sbjct 10 KSWHPLLMRNQEKVWKDEQAHKEEMKRVEQLRREIEEERQLLELHR 55
> 7295920
Length=521
Score = 32.3 bits (72), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 23/31 (74%), Gaps = 0/31 (0%)
Query 10 KPFHPGNYQNQEKVWLAEQKLKEEQKKEKEL 40
K +HP +NQE+VW AE++ K E++K ++L
Sbjct 10 KSWHPHTMKNQERVWKAEEQAKMEERKLQDL 40
> Hs8923271
Length=425
Score = 30.0 bits (66), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 15/27 (55%), Positives = 19/27 (70%), Gaps = 0/27 (0%)
Query 10 KPFHPGNYQNQEKVWLAEQKLKEEQKK 36
K +HP +N EKVW AEQK + E+KK
Sbjct 10 KSWHPQTLRNVEKVWKAEQKHEAERKK 36
> Hs5453642
Length=1233
Score = 29.6 bits (65), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 31/100 (31%), Positives = 48/100 (48%), Gaps = 11/100 (11%)
Query 50 IEELRRALRGEAKAALQLQKKEEAVSPAEELRRKAKEAARLQALQRKHQEAQNRLKKLSK 109
+EE+R+ L G K LQK+ A+ E +R + LQA K Q+ + L K +
Sbjct 891 MEEIRKKLGGANKEMTHLQKEVTAIETKLEQKRSDRHNL-LQAC--KMQDIKLPLSKGTM 947
Query 110 STLYDEDAFTQGHTSVFG-----SLYSKE---ENKWGYRC 141
+ E+ +QG SV G S+Y++E E +G C
Sbjct 948 DDISQEEGSSQGEDSVSGSQRISSIYAREALIEIDYGDLC 987
> CE26143
Length=646
Score = 28.1 bits (61), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 41/73 (56%), Gaps = 5/73 (6%)
Query 13 HPGNYQNQEKVWLA--EQKLKEEQKKEKELEERRREEVKIEELRRALRGEAKAALQLQKK 70
+PG+ +NQE+ W A +++ E +KK K LEER V EE R ++ + +L+K
Sbjct 364 NPGDDENQEEAWEAKMQEREVEMEKKRKILEERVNSAVNDEETHRLVKEMMENEAELKK- 422
Query 71 EEAVSPAEELRRK 83
A S E+LR K
Sbjct 423 --ARSEHEKLRSK 433
Lambda K H
0.308 0.123 0.340
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3094507556
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40