bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1020_orf1
Length=154
Score E
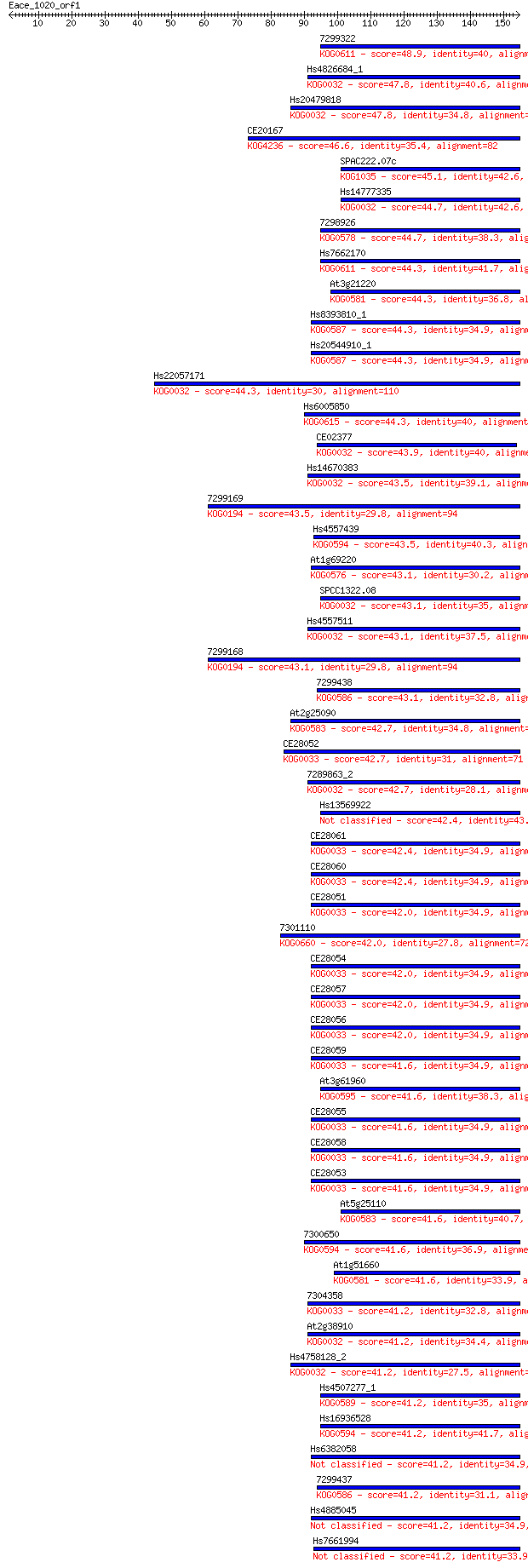
Sequences producing significant alignments: (Bits) Value
7299322 48.9 4e-06
Hs4826684_1 47.8 8e-06
Hs20479818 47.8 9e-06
CE20167 46.6 2e-05
SPAC222.07c 45.1 6e-05
Hs14777335 44.7 7e-05
7298926 44.7 7e-05
Hs7662170 44.3 8e-05
At3g21220 44.3 8e-05
Hs8393810_1 44.3 9e-05
Hs20544910_1 44.3 9e-05
Hs22057171 44.3 1e-04
Hs6005850 44.3 1e-04
CE02377 43.9 1e-04
Hs14670383 43.5 2e-04
7299169 43.5 2e-04
Hs4557439 43.5 2e-04
At1g69220 43.1 2e-04
SPCC1322.08 43.1 2e-04
Hs4557511 43.1 2e-04
7299168 43.1 2e-04
7299438 43.1 2e-04
At2g25090 42.7 2e-04
CE28052 42.7 2e-04
7289863_2 42.7 2e-04
Hs13569922 42.4 3e-04
CE28061 42.4 3e-04
CE28060 42.4 4e-04
CE28051 42.0 4e-04
7301110 42.0 4e-04
CE28054 42.0 4e-04
CE28057 42.0 5e-04
CE28056 42.0 5e-04
CE28059 41.6 5e-04
At3g61960 41.6 5e-04
CE28055 41.6 5e-04
CE28058 41.6 6e-04
CE28053 41.6 6e-04
At5g25110 41.6 6e-04
7300650 41.6 7e-04
At1g51660 41.6 7e-04
7304358 41.2 7e-04
At2g38910 41.2 7e-04
Hs4758128_2 41.2 7e-04
Hs4507277_1 41.2 7e-04
Hs16936528 41.2 8e-04
Hs6382058 41.2 8e-04
7299437 41.2 8e-04
Hs4885045 41.2 8e-04
Hs7661994 41.2 9e-04
> 7299322
Length=434
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 24/62 (38%), Positives = 39/62 (62%), Gaps = 2/62 (3%)
Query 95 FTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQVPSE--QIEAEIEVLKSLDHPN 152
F +KKLG G++G V I K +G E IKTI + + + ++ +I E++++ S+ HPN
Sbjct 82 FDIIKKLGQGTYGKVQLGINKETGQEVAIKTIKKCKIEAEADLVRIRREVQIMSSVHHPN 141
Query 153 II 154
II
Sbjct 142 II 143
> Hs4826684_1
Length=370
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 40/69 (57%), Gaps = 5/69 (7%)
Query 91 ITDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQ-----VPSEQIEAEIEVL 145
+ D + ++LGSG F V + EKS+GL+ K I + R++ V E IE E+ +L
Sbjct 9 VDDYYDTGEELGSGQFAVVKKCREKSTGLQYAAKFIKKRRTKSSRRGVSREDIEREVSIL 68
Query 146 KSLDHPNII 154
K + HPN+I
Sbjct 69 KEIQHPNVI 77
> Hs20479818
Length=514
Score = 47.8 bits (112), Expect = 9e-06, Method: Composition-based stats.
Identities = 24/70 (34%), Positives = 44/70 (62%), Gaps = 1/70 (1%)
Query 86 QTGLAITDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQVPSEQ-IEAEIEV 144
+ G AI +++TF + LG GSFG V +K + + IK +N++++ + + +E E+ +
Sbjct 107 ENGAAIEEIYTFGRILGKGSFGIVIEATDKETETKWAIKKVNKEKAGSSAVKLLEREVNI 166
Query 145 LKSLDHPNII 154
LKS+ H +II
Sbjct 167 LKSVKHEHII 176
> CE20167
Length=722
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 48/89 (53%), Gaps = 7/89 (7%)
Query 73 VRRSLFVGRRNL--AQTGLAITDLFTFV--KKLGSGSFGDVHRVIEKSSGLERVIKTINR 128
++ SL RN A+ L +L+ + K LGSG FG V+ I++ SG E +K I++
Sbjct 400 IKESLQPPSRNEDNAEQALEFANLYQVLSDKTLGSGQFGTVYSAIQRHSGKEVAVKVISK 459
Query 129 DR---SQVPSEQIEAEIEVLKSLDHPNII 154
+R +E + AE+ +L+ HP I+
Sbjct 460 ERFSKKGSGAESMRAEVAILQQTCHPGIV 488
> SPAC222.07c
Length=639
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 23/54 (42%), Positives = 30/54 (55%), Gaps = 0/54 (0%)
Query 101 LGSGSFGDVHRVIEKSSGLERVIKTINRDRSQVPSEQIEAEIEVLKSLDHPNII 154
LG G FG V+ V K G E +K IN Q+ +I EI+ L +DHPN+I
Sbjct 177 LGRGGFGSVYHVRNKIDGAEYAMKKINSTFQQMSYSKIFREIKCLAKMDHPNVI 230
> Hs14777335
Length=795
Score = 44.7 bits (104), Expect = 7e-05, Method: Composition-based stats.
Identities = 23/54 (42%), Positives = 31/54 (57%), Gaps = 1/54 (1%)
Query 101 LGSGSFGDVHRVIEKSSGLERVIKTINRDRSQVPSEQIEAEIEVLKSLDHPNII 154
LG G FG VHR EKS+GL K I + +S E ++ EI ++ L H N+I
Sbjct 497 LGGGRFGQVHRCTEKSTGLPLAAKII-KVKSAKDREDVKNEINIMNQLSHVNLI 549
> 7298926
Length=704
Score = 44.7 bits (104), Expect = 7e-05, Method: Composition-based stats.
Identities = 23/60 (38%), Positives = 37/60 (61%), Gaps = 1/60 (1%)
Query 95 FTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQVPSEQIEAEIEVLKSLDHPNII 154
+T ++K+G G+ G V+ IE S+G+E IK +N + Q E I EI V++ HPN++
Sbjct 430 YTKMEKIGQGASGTVYTAIESSTGMEVAIKQMNLSQ-QPKKELIINEILVMRENKHPNVV 488
> Hs7662170
Length=661
Score = 44.3 bits (103), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 25/64 (39%), Positives = 38/64 (59%), Gaps = 6/64 (9%)
Query 95 FTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQVPSEQ----IEAEIEVLKSLDH 150
+ + LG G++G V R E+ SG IK+I +D+ + EQ I EIE++ SL+H
Sbjct 55 YELQETLGKGTYGKVKRATERFSGRVVAIKSIRKDK--IKDEQDMVHIRREIEIMSSLNH 112
Query 151 PNII 154
P+II
Sbjct 113 PHII 116
> At3g21220
Length=348
Score = 44.3 bits (103), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 21/57 (36%), Positives = 34/57 (59%), Gaps = 0/57 (0%)
Query 98 VKKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQVPSEQIEAEIEVLKSLDHPNII 154
V ++GSG+ G V++VI + +K I + QI EIE+L+S+DHPN++
Sbjct 73 VNRIGSGAGGTVYKVIHTPTSRPFALKVIYGNHEDTVRRQICREIEILRSVDHPNVV 129
> Hs8393810_1
Length=336
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 39/64 (60%), Gaps = 3/64 (4%)
Query 92 TDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQVPSEQIEAEIEVLKSL-DH 150
+D + + +G G++G V +V+ K +G + +K + D E+IEAE +LK+L DH
Sbjct 18 SDTWEITETIGKGTYGKVFKVLNKKNGQKAAVKIL--DPIHDIDEEIEAEYNILKALSDH 75
Query 151 PNII 154
PN++
Sbjct 76 PNVV 79
> Hs20544910_1
Length=336
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 39/64 (60%), Gaps = 3/64 (4%)
Query 92 TDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQVPSEQIEAEIEVLKSL-DH 150
+D + + +G G++G V +V+ K +G + +K + D E+IEAE +LK+L DH
Sbjct 18 SDTWEITETIGKGTYGKVFKVLNKKNGQKAAVKIL--DPIHDIDEEIEAEYNILKALSDH 75
Query 151 PNII 154
PN++
Sbjct 76 PNVV 79
> Hs22057171
Length=456
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 50/110 (45%), Gaps = 8/110 (7%)
Query 45 LELEDFCRMYWEILCRVREKYYPEKAMRVRRSLFVGRRNLAQTGLAITDLFTFVKKLGSG 104
L L D + I C +R + + E + + + TG T L F G G
Sbjct 131 LSLPDLADL---IECPMRLRGWAEGETGIPQHELAPWKPELTTGPHATPLPLF----GRG 183
Query 105 SFGDVHRVIEKSSGLERVIKTINRDRSQVPSEQIEAEIEVLKSLDHPNII 154
FG VH+ E ++GL+ K I + R E+++ EI V+ LDH N+I
Sbjct 184 RFGQVHKCEETATGLKLAAKII-KTRGMKDKEEVKNEISVMNQLDHANLI 232
> Hs6005850
Length=543
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/72 (36%), Positives = 39/72 (54%), Gaps = 7/72 (9%)
Query 90 AITDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINR-------DRSQVPSEQIEAEI 142
A+ D + K LGSG+ G+V E+ + + IK I++ R P+ +E EI
Sbjct 215 ALRDEYIMSKTLGSGACGEVKLAFERKTCKKVAIKIISKRKFAIGSAREADPALNVETEI 274
Query 143 EVLKSLDHPNII 154
E+LK L+HP II
Sbjct 275 EILKKLNHPCII 286
> CE02377
Length=1211
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 24/60 (40%), Positives = 33/60 (55%), Gaps = 1/60 (1%)
Query 94 LFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQVPSEQIEAEIEVLKSLDHPNI 153
L+ K LG G FG V+ VIEK +G E K I + R + ++E E+ +L L HP I
Sbjct 44 LYQVTKLLGDGKFGKVYCVIEKETGKEFAAKFI-KIRKEADRAEVEREVSILTQLRHPRI 102
> Hs14670383
Length=370
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 40/69 (57%), Gaps = 5/69 (7%)
Query 91 ITDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQ-----VPSEQIEAEIEVL 145
+ D + ++LGSG F V + EKS+GLE K I + +S+ V E+IE E+ +L
Sbjct 19 VEDFYDIGEELGSGQFAIVKKCREKSTGLEYAAKFIKKRQSRASRRGVSREEIEREVSIL 78
Query 146 KSLDHPNII 154
+ + H N+I
Sbjct 79 RQVLHHNVI 87
> 7299169
Length=592
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 48/97 (49%), Gaps = 6/97 (6%)
Query 61 VREKYYPEKAMRVRRSLFVGRRNLAQTGLAITDLFTFVKKLGSGSFGDVHRVIEKSSGLE 120
+ +Y+ E + V+ + R + D ++++G G+FGDV++ KS+ L+
Sbjct 296 IMHQYHSELPVTVKSGAILRRPVCRERWELSNDDVVLLERIGRGNFGDVYKAKLKSTKLD 355
Query 121 RVIKTINRDRSQVPSEQIE---AEIEVLKSLDHPNII 154
+KT R +P EQ E +LK DHPNI+
Sbjct 356 VAVKTC---RMTLPDEQKRKFLQEGRILKQYDHPNIV 389
> Hs4557439
Length=305
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/64 (39%), Positives = 37/64 (57%), Gaps = 3/64 (4%)
Query 93 DLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQ--VPSEQIEAEIEVLKSLDH 150
D+F V+K+G G++G V++ + +G +K I D VPS I EI +LK L H
Sbjct 2 DMFQKVEKIGEGTYGVVYKAKNRETGQLVALKKIRLDLEMEGVPSTAIR-EISLLKELKH 60
Query 151 PNII 154
PNI+
Sbjct 61 PNIV 64
> At1g69220
Length=836
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 36/63 (57%), Gaps = 0/63 (0%)
Query 92 TDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQVPSEQIEAEIEVLKSLDHP 151
T + F+ +LG GS+G V++ + + +K I+ + E+I EIE+L+ +HP
Sbjct 246 TTKYEFLNELGKGSYGSVYKARDLKTSEIVAVKVISLTEGEEGYEEIRGEIEMLQQCNHP 305
Query 152 NII 154
N++
Sbjct 306 NVV 308
> SPCC1322.08
Length=580
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/71 (29%), Positives = 38/71 (53%), Gaps = 11/71 (15%)
Query 95 FTFVKKLGSGSFGDVHRVIEKSSGLERVIKTIN-----------RDRSQVPSEQIEAEIE 143
+T ++K+G G+F +V++ I +G + IK + R R V S I E++
Sbjct 124 YTLLQKMGDGAFSNVYKAIHNRTGEKVAIKVVQRAQPNTDPRDPRKRQGVESHNILKEVQ 183
Query 144 VLKSLDHPNII 154
+++ + HPNII
Sbjct 184 IMRRVKHPNII 194
> Hs4557511
Length=454
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/69 (34%), Positives = 37/69 (53%), Gaps = 5/69 (7%)
Query 91 ITDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINR-----DRSQVPSEQIEAEIEVL 145
+ D + ++LGSG F V + +K +G E K I + R V E+IE E+ +L
Sbjct 9 VEDHYEMGEELGSGQFAIVRKCRQKGTGKEYAAKFIKKRRLSSSRRGVSREEIEREVNIL 68
Query 146 KSLDHPNII 154
+ + HPNII
Sbjct 69 REIRHPNII 77
> 7299168
Length=804
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 48/97 (49%), Gaps = 6/97 (6%)
Query 61 VREKYYPEKAMRVRRSLFVGRRNLAQTGLAITDLFTFVKKLGSGSFGDVHRVIEKSSGLE 120
+ +Y+ E + V+ + R + D ++++G G+FGDV++ KS+ L+
Sbjct 508 IMHQYHSELPVTVKSGAILRRPVCRERWELSNDDVVLLERIGRGNFGDVYKAKLKSTKLD 567
Query 121 RVIKTINRDRSQVPSEQIE---AEIEVLKSLDHPNII 154
+KT R +P EQ E +LK DHPNI+
Sbjct 568 VAVKTC---RMTLPDEQKRKFLQEGRILKQYDHPNIV 601
> 7299438
Length=705
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 36/62 (58%), Gaps = 1/62 (1%)
Query 94 LFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQVPSEQ-IEAEIEVLKSLDHPN 152
++ +K LG G+F V I +G E IK I++ + + Q + E++++K L+HPN
Sbjct 97 VYKIIKTLGKGNFAKVKLAIHVPTGREVAIKVIDKTQLNTSARQKLYREVKIMKLLNHPN 156
Query 153 II 154
I+
Sbjct 157 IV 158
> At2g25090
Length=469
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/72 (33%), Positives = 41/72 (56%), Gaps = 3/72 (4%)
Query 86 QTGLAITDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRD---RSQVPSEQIEAEI 142
++ + D + + LG+G+F V+ E S+G + IK I +D + + EQIE EI
Sbjct 6 RSSTVLFDKYNIGRLLGTGNFAKVYHGTEISTGDDVAIKVIKKDHVFKRRGMMEQIEREI 65
Query 143 EVLKSLDHPNII 154
V++ L HPN++
Sbjct 66 AVMRLLRHPNVV 77
> CE28052
Length=137
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 38/72 (52%), Gaps = 1/72 (1%)
Query 84 LAQTGLAITDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDR-SQVPSEQIEAEI 142
+ +D + ++LG G+F V R + K++GLE K IN + S +++E E
Sbjct 1 MMNASTKFSDNYDVKEELGKGAFSVVRRCVHKTTGLEFAAKIINTKKLSARDFQKLEREA 60
Query 143 EVLKSLDHPNII 154
+ + L HPNI+
Sbjct 61 RICRKLQHPNIV 72
> 7289863_2
Length=93
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 18/64 (28%), Positives = 39/64 (60%), Gaps = 0/64 (0%)
Query 91 ITDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQVPSEQIEAEIEVLKSLDH 150
I + ++ + +G G+F V ++ + +G +K I++++ + I+AE+ V+K L+H
Sbjct 6 IRNTYSLGRIIGDGNFAIVFKIKHRQTGHSYALKIIDKNKCKGKEHYIDAEVRVMKKLNH 65
Query 151 PNII 154
P+II
Sbjct 66 PHII 69
> Hs13569922
Length=628
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 40/64 (62%), Gaps = 7/64 (10%)
Query 95 FTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQVPSEQ----IEAEIEVLKSLDH 150
+ F++ LG G++G V + E SSG IK+I +D+ + EQ I EIE++ SL+H
Sbjct 53 YEFLETLGKGTYGKVKKARE-SSGRLVAIKSIRKDK--IKDEQDLMHIRREIEIMSSLNH 109
Query 151 PNII 154
P+II
Sbjct 110 PHII 113
> CE28061
Length=302
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 37/64 (57%), Gaps = 1/64 (1%)
Query 92 TDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDR-SQVPSEQIEAEIEVLKSLDH 150
+D + ++LG G+F V R + K++GLE K IN + S +++E E + + L H
Sbjct 9 SDNYDVKEELGKGAFSVVRRCVHKTTGLEFAAKIINTKKLSARDFQKLEREARICRKLQH 68
Query 151 PNII 154
PNI+
Sbjct 69 PNIV 72
> CE28060
Length=302
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 37/64 (57%), Gaps = 1/64 (1%)
Query 92 TDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDR-SQVPSEQIEAEIEVLKSLDH 150
+D + ++LG G+F V R + K++GLE K IN + S +++E E + + L H
Sbjct 9 SDNYDVKEELGKGAFSVVRRCVHKTTGLEFAAKIINTKKLSARDFQKLEREARICRKLQH 68
Query 151 PNII 154
PNI+
Sbjct 69 PNIV 72
> CE28051
Length=559
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 37/64 (57%), Gaps = 1/64 (1%)
Query 92 TDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDR-SQVPSEQIEAEIEVLKSLDH 150
+D + ++LG G+F V R + K++GLE K IN + S +++E E + + L H
Sbjct 9 SDNYDVKEELGKGAFSVVRRCVHKTTGLEFAAKIINTKKLSARDFQKLEREARICRKLQH 68
Query 151 PNII 154
PNI+
Sbjct 69 PNIV 72
> 7301110
Length=366
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 20/73 (27%), Positives = 44/73 (60%), Gaps = 1/73 (1%)
Query 83 NLAQTGLAITDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRD-RSQVPSEQIEAE 141
++ +T I D++ ++ +GSG++G V + + + + + IK + R +S V +++ E
Sbjct 13 DINRTEWEIPDIYQDLQPVGSGAYGQVSKAVVRGTNMHVAIKKLARPFQSAVHAKRTYRE 72
Query 142 IEVLKSLDHPNII 154
+ +LK +DH N+I
Sbjct 73 LRLLKHMDHENVI 85
> CE28054
Length=482
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 37/64 (57%), Gaps = 1/64 (1%)
Query 92 TDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDR-SQVPSEQIEAEIEVLKSLDH 150
+D + ++LG G+F V R + K++GLE K IN + S +++E E + + L H
Sbjct 9 SDNYDVKEELGKGAFSVVRRCVHKTTGLEFAAKIINTKKLSARDFQKLEREARICRKLQH 68
Query 151 PNII 154
PNI+
Sbjct 69 PNIV 72
> CE28057
Length=520
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 37/64 (57%), Gaps = 1/64 (1%)
Query 92 TDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDR-SQVPSEQIEAEIEVLKSLDH 150
+D + ++LG G+F V R + K++GLE K IN + S +++E E + + L H
Sbjct 9 SDNYDVKEELGKGAFSVVRRCVHKTTGLEFAAKIINTKKLSARDFQKLEREARICRKLQH 68
Query 151 PNII 154
PNI+
Sbjct 69 PNIV 72
> CE28056
Length=518
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 37/64 (57%), Gaps = 1/64 (1%)
Query 92 TDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDR-SQVPSEQIEAEIEVLKSLDH 150
+D + ++LG G+F V R + K++GLE K IN + S +++E E + + L H
Sbjct 9 SDNYDVKEELGKGAFSVVRRCVHKTTGLEFAAKIINTKKLSARDFQKLEREARICRKLQH 68
Query 151 PNII 154
PNI+
Sbjct 69 PNIV 72
> CE28059
Length=350
Score = 41.6 bits (96), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 37/64 (57%), Gaps = 1/64 (1%)
Query 92 TDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDR-SQVPSEQIEAEIEVLKSLDH 150
+D + ++LG G+F V R + K++GLE K IN + S +++E E + + L H
Sbjct 9 SDNYDVKEELGKGAFSVVRRCVHKTTGLEFAAKIINTKKLSARDFQKLEREARICRKLQH 68
Query 151 PNII 154
PNI+
Sbjct 69 PNIV 72
> At3g61960
Length=648
Score = 41.6 bits (96), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 23/61 (37%), Positives = 35/61 (57%), Gaps = 1/61 (1%)
Query 95 FTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRD-RSQVPSEQIEAEIEVLKSLDHPNI 153
+ ++GSGSF V +SSGLE +K I++ S + + EI +L ++DHPNI
Sbjct 10 YALGPRIGSGSFAVVWLAKHRSSGLEVAVKEIDKKLLSPKVRDNLLKEISILSTIDHPNI 69
Query 154 I 154
I
Sbjct 70 I 70
> CE28055
Length=533
Score = 41.6 bits (96), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 37/64 (57%), Gaps = 1/64 (1%)
Query 92 TDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDR-SQVPSEQIEAEIEVLKSLDH 150
+D + ++LG G+F V R + K++GLE K IN + S +++E E + + L H
Sbjct 9 SDNYDVKEELGKGAFSVVRRCVHKTTGLEFAAKIINTKKLSARDFQKLEREARICRKLQH 68
Query 151 PNII 154
PNI+
Sbjct 69 PNIV 72
> CE28058
Length=571
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 37/64 (57%), Gaps = 1/64 (1%)
Query 92 TDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDR-SQVPSEQIEAEIEVLKSLDH 150
+D + ++LG G+F V R + K++GLE K IN + S +++E E + + L H
Sbjct 9 SDNYDVKEELGKGAFSVVRRCVHKTTGLEFAAKIINTKKLSARDFQKLEREARICRKLQH 68
Query 151 PNII 154
PNI+
Sbjct 69 PNIV 72
> CE28053
Length=720
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 37/64 (57%), Gaps = 1/64 (1%)
Query 92 TDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDR-SQVPSEQIEAEIEVLKSLDH 150
+D + ++LG G+F V R + K++GLE K IN + S +++E E + + L H
Sbjct 9 SDNYDVKEELGKGAFSVVRRCVHKTTGLEFAAKIINTKKLSARDFQKLEREARICRKLQH 68
Query 151 PNII 154
PNI+
Sbjct 69 PNIV 72
> At5g25110
Length=460
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 22/56 (39%), Positives = 35/56 (62%), Gaps = 2/56 (3%)
Query 101 LGSGSFGDVHRVIEKSSGLERVIKTINRDRSQVPS--EQIEAEIEVLKSLDHPNII 154
LG G+FG V+ E ++G IK IN+D+ + EQI+ EI +++ + HPNI+
Sbjct 21 LGKGTFGKVYYGKEITTGESVAIKIINKDQVKREGMMEQIKREISIMRLVRHPNIV 76
> 7300650
Length=314
Score = 41.6 bits (96), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 38/67 (56%), Gaps = 3/67 (4%)
Query 90 AITDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTI--NRDRSQVPSEQIEAEIEVLKS 147
I D F +K+G G++G V++ S+G + +K I + VPS I EI +LK+
Sbjct 3 TILDNFQRAEKIGEGTYGIVYKARSNSTGQDVALKKIRLEGETEGVPSTAIR-EISLLKN 61
Query 148 LDHPNII 154
L HPN++
Sbjct 62 LKHPNVV 68
> At1g51660
Length=366
Score = 41.6 bits (96), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 34/56 (60%), Gaps = 0/56 (0%)
Query 99 KKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQVPSEQIEAEIEVLKSLDHPNII 154
++GSG+ G V++VI + S +K I + + QI EIE+L+ ++HPN++
Sbjct 83 NRIGSGAGGTVYKVIHRPSSRLYALKVIYGNHEETVRRQICREIEILRDVNHPNVV 138
> 7304358
Length=493
Score = 41.2 bits (95), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 37/65 (56%), Gaps = 1/65 (1%)
Query 91 ITDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDR-SQVPSEQIEAEIEVLKSLD 149
+D + ++LG G+F V R ++KS+G E K IN + + +++E E + + L
Sbjct 10 FSDNYDIKEELGKGAFSIVKRCVQKSTGFEFAAKIINTKKLTARDFQKLEREARICRKLH 69
Query 150 HPNII 154
HPNI+
Sbjct 70 HPNIV 74
> At2g38910
Length=583
Score = 41.2 bits (95), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 37/67 (55%), Gaps = 3/67 (4%)
Query 91 ITDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQVPS--EQIEAEIEVLKSL 148
+ D+++ +KLG G FG ++K +G E KTI + + P E + EI+++ L
Sbjct 130 LKDIYSVGRKLGQGQFGTTFLCVDKKTGKEFACKTIAKRKLTTPEDVEDVRREIQIMHHL 189
Query 149 D-HPNII 154
HPN+I
Sbjct 190 SGHPNVI 196
> Hs4758128_2
Length=385
Score = 41.2 bits (95), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 19/69 (27%), Positives = 37/69 (53%), Gaps = 0/69 (0%)
Query 86 QTGLAITDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQVPSEQIEAEIEVL 145
Q IT+ + + +G G+F V +E+S+ E +K I + + + I+ E+ +L
Sbjct 37 QIPATITERYKVGRTIGDGNFAVVKECVERSTAREYALKIIKKSKCRGKEHMIQNEVSIL 96
Query 146 KSLDHPNII 154
+ + HPNI+
Sbjct 97 RRVKHPNIV 105
> Hs4507277_1
Length=292
Score = 41.2 bits (95), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 21/61 (34%), Positives = 35/61 (57%), Gaps = 1/61 (1%)
Query 95 FTFVKKLGSGSFGDVHRVIEKSSGLERVIKTIN-RDRSQVPSEQIEAEIEVLKSLDHPNI 153
+ +++ +G GS+G+V V + G + VIK +N R+ S E E ++L L HPNI
Sbjct 6 YCYLRVVGKGSYGEVTLVKHRRDGKQYVIKKLNLRNASSRERRAAEQEAQLLSQLKHPNI 65
Query 154 I 154
+
Sbjct 66 V 66
> Hs16936528
Length=298
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 36/62 (58%), Gaps = 3/62 (4%)
Query 95 FTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQ--VPSEQIEAEIEVLKSLDHPN 152
F V+K+G G++G V++ K +G +K I D VPS I EI +LK L+HPN
Sbjct 4 FQKVEKIGEGTYGVVYKARNKLTGEVVALKKIRLDTETEGVPSTAIR-EISLLKELNHPN 62
Query 153 II 154
I+
Sbjct 63 IV 64
> Hs6382058
Length=1148
Score = 41.2 bits (95), Expect = 8e-04, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 35/63 (55%), Gaps = 3/63 (4%)
Query 92 TDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQVPSEQIEAEIEVLKSLDHP 151
TD+ T KLG G +G+V+ + K L +KT+ D +V E+ E V+K + HP
Sbjct 258 TDI-TMKHKLGGGQYGEVYEGVWKKYSLTVAVKTLKEDTMEV--EEFLKEAAVMKEIKHP 314
Query 152 NII 154
N++
Sbjct 315 NLV 317
> 7299437
Length=604
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 36/62 (58%), Gaps = 1/62 (1%)
Query 94 LFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDR-SQVPSEQIEAEIEVLKSLDHPN 152
++ +K LG G+F V I +G E IK I++ + + +++ E+ ++K L+HPN
Sbjct 62 VYKIIKTLGKGNFAKVKLAIHLPTGREVAIKLIDKTALNTIARQKLYREVNIMKKLNHPN 121
Query 153 II 154
I+
Sbjct 122 IV 123
> Hs4885045
Length=1130
Score = 41.2 bits (95), Expect = 8e-04, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 35/63 (55%), Gaps = 3/63 (4%)
Query 92 TDLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQVPSEQIEAEIEVLKSLDHP 151
TD+ T KLG G +G+V+ + K L +KT+ D +V E+ E V+K + HP
Sbjct 240 TDI-TMKHKLGGGQYGEVYEGVWKKYSLTVAVKTLKEDTMEV--EEFLKEAAVMKEIKHP 296
Query 152 NII 154
N++
Sbjct 297 NLV 299
> Hs7661994
Length=1152
Score = 41.2 bits (95), Expect = 9e-04, Method: Composition-based stats.
Identities = 21/62 (33%), Positives = 35/62 (56%), Gaps = 1/62 (1%)
Query 93 DLFTFVKKLGSGSFGDVHRVIEKSSGLERVIKTINRDRSQVPSEQIEAEIEVLKSLDHPN 152
D + + +LG G+FG V++ K + + K I+ +S+ E EI++L S DHPN
Sbjct 32 DFWEIIGELGDGAFGKVYKAQNKETSVLAAAKVID-TKSEEELEDYMVEIDILASCDHPN 90
Query 153 II 154
I+
Sbjct 91 IV 92
Lambda K H
0.325 0.141 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1997677330
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40