bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1007_orf6
Length=376
Score E
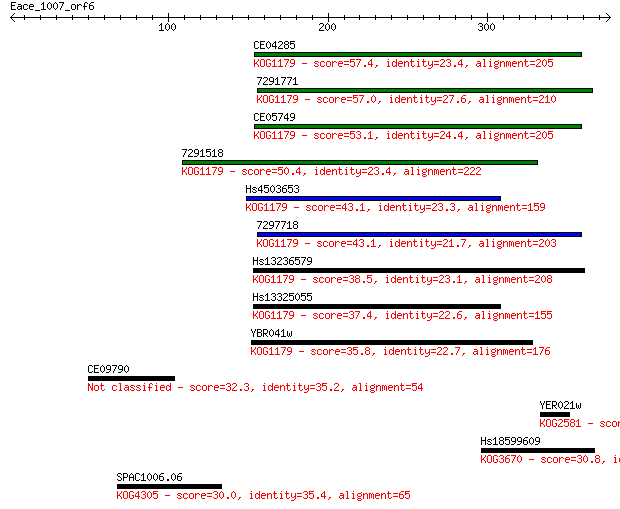
Sequences producing significant alignments: (Bits) Value
CE04285 57.4 5e-08
7291771 57.0 7e-08
CE05749 53.1 9e-07
7291518 50.4 7e-06
Hs4503653 43.1 0.001
7297718 43.1 0.001
Hs13236579 38.5 0.024
Hs13325055 37.4 0.049
YBR041w 35.8 0.14
CE09790 32.3 1.6
YER021w 31.6 3.2
Hs18599609 30.8 5.2
SPAC1006.06 30.0 9.6
> CE04285
Length=655
Score = 57.4 bits (137), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 48/207 (23%), Positives = 89/207 (42%), Gaps = 15/207 (7%)
Query 154 CCLYTADLEGHLRATWVSNSRFLSAGFCWLEAVNLTESDRLLLGVQLSHEV-GLHALAAA 212
C +YT+ G+ + + + R+ +A + +SD + + + + H G+ + +
Sbjct 247 CYIYTSGTTGNPKPAVIKHFRYFWIAMGAGKAFGINKSDVVYITMPMYHSAAGIMGIGSL 306
Query 213 IACKGALLLRSRSSLLSLWPDVKAMDATILWHTGMMWCRLLKAHESYQGGDQTQRLGSWS 272
IA ++R + S + W D + T + G + LL A+ + RL W
Sbjct 307 IAFGSTAVIRKKFSASNFWKDCVKYNVTATQYIGEICRYLLAANPCPEEKQHNVRL-MW- 364
Query 273 GNPQLRASIGTGLAGALWPVVKKVFNIPRILEFYSPRNLQTPQILFNAWDMLGACAFVPD 332
G GL G +W F I +I E Y + + + N + +GAC F+P
Sbjct 365 ---------GNGLRGQIWKEFVGRFGIKKIGELYG--STEGNSNIVNVDNHVGACGFMPI 413
Query 333 FA-WNSKELERIVEFNEKTGEVYREKN 358
+ S R+++ + TGE+ R+KN
Sbjct 414 YPHIGSLYPVRLIKVDRATGELERDKN 440
> 7291771
Length=661
Score = 57.0 bits (136), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 58/218 (26%), Positives = 95/218 (43%), Gaps = 24/218 (11%)
Query 156 LYTADLEGHLRATWVSNSRFL--SAGFCWLEAVNLTESDRLLLGVQLSHEVG-LHALAAA 212
+YT+ G +A ++N RFL SAG ++ + ++ D + + L H G + + A
Sbjct 263 VYTSGTTGLPKAAVITNLRFLFMSAGSYYM--LKMSSDDVVYDPLPLYHTAGGIVGVGNA 320
Query 213 IACKGALLLRSRSSLLSLWPDVKAMDATILWHTGMMWCRLLKAHESYQGGDQTQRLGSWS 272
I ++LR + S + W D + T+ + G + CR L A SY Q
Sbjct 321 ILNGSTVVLRKKFSARNFWLDCDRHNCTVAQYIGEL-CRYLLA-TSYSPDQQKH------ 372
Query 273 GNPQLRASIGTGLAGALWPVVKKVFNIPRILEFYSPRNLQTPQILFNAWDMLGACAFVPD 332
LR G GL +W + F IP I E Y + L N + +GA FVP
Sbjct 373 ---NLRLMYGNGLRPQIWSQFVRRFGIPHIGEIYGATEGNSN--LINITNRVGAIGFVPV 427
Query 333 FAWNSKELERIVEFNEKTGEVYREKNN-----QEGQGG 365
+ + ++ ++ +E TGE+ ++ Q GQ G
Sbjct 428 YGSSLYPVQ-VLRCDEYTGELLKDSKGHCIRCQPGQAG 464
> CE05749
Length=650
Score = 53.1 bits (126), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 50/208 (24%), Positives = 91/208 (43%), Gaps = 18/208 (8%)
Query 154 CCLYTADLEGHLRATWVSNSRFLSAGFCWLEAVNLTESDRLLLGVQLSHE-VGLHALAAA 212
C +YT+ G +A + + R+ S ++ + SDR+ + + + H G+ + A
Sbjct 244 CFIYTSGTTGMPKAAVMKHFRYYSIAVGAAKSFGIRPSDRMYVSMPIYHTAAGILGVGQA 303
Query 213 IACKGALLLRSRSSLLSLWPDVKAMDATILWHTGMMWCRLLKAHESYQGGDQTQRLGSWS 272
+ + ++R + S + W D D T+ + G + CR L A Q +
Sbjct 304 LLGGSSCVIRKKFSASNFWRDCVKYDCTVSQYIGEI-CRYLLA----------QPVVEEE 352
Query 273 GNPQLRASIGTGLAGALW-PVVKKVFNIPRILEFYSPRNLQTPQILFNAWDMLGACAFVP 331
++R +G GL +W P V + F + RI E Y + + L N +GAC F+P
Sbjct 353 SRHRMRLLVGNGLRAEIWQPFVDR-FRV-RIGELYG--STEGTSSLVNIDGHVGACGFLP 408
Query 332 DFAWNSK-ELERIVEFNEKTGEVYREKN 358
K R+++ ++ TGE R +
Sbjct 409 ISPLTKKMHPVRLIKVDDVTGEAIRTSD 436
> 7291518
Length=664
Score = 50.4 bits (119), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 52/227 (22%), Positives = 92/227 (40%), Gaps = 20/227 (8%)
Query 109 NDE--QHKCKPSGFPMLLKEPLRKLVGSHSERQAASGAHCAEMSLPCCCLYTADLEGHLR 166
NDE Q G L + L L+ + ++ + A+GA A+ +YT+ G +
Sbjct 224 NDESNQEVVASEGLSQGLAQQLNGLLETAAKDKVAAGASRADHHDKLVYIYTSGTTGLPK 283
Query 167 ATWVSNSR--FLSAGFCWLEAVNLTESDRLLLGVQLSHEVG-LHALAAAIACKGALLLRS 223
A +++SR F++AG + + + D + L H G + ++ A+ +++R
Sbjct 284 AAVITHSRYFFIAAGIHY--TLGFKDQDVFYTPLPLYHTAGGVMSMGQALLFGSTVVIRK 341
Query 224 RSSLLSLWPDVKAMDATILWHTGMMWCRLLKAHESYQGGDQTQRLGSWSGNPQLRASIGT 283
+ S + D T+ + G M R + A S N Q+R G
Sbjct 342 KFSASGYFSDCARFQCTVGQYIGEM-ARYILATPS----------APHDRNHQVRMVFGN 390
Query 284 GLAGALWPVVKKVFNIPRILEFYSPRNLQTPQILFNAWDMLGACAFV 330
GL +WP + F I ++ EFY + N +GA F+
Sbjct 391 GLRPQIWPQFVERFGIRKVGEFYGATEGNAN--IMNNDSTVGAIGFI 435
> Hs4503653
Length=620
Score = 43.1 bits (100), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 37/164 (22%), Positives = 64/164 (39%), Gaps = 21/164 (12%)
Query 149 MSLPCCCLYTADLEGHLRATWVSNSRF-LSAGFCWLEAVNLTESDRLLLGVQLSHE---- 203
S P +YT+ G +A +++ R G ++ L D + + + H
Sbjct 216 FSTPALYIYTSGTTGLPKAAMITHQRIWYGTGLTFVSG--LKADDVIYITLPFYHSAALL 273
Query 204 VGLHALAAAIACKGALLLRSRSSLLSLWPDVKAMDATILWHTGMMWCRLLKAHESYQGGD 263
+G+H I L LR++ S W D + + T++ + G + L + + D
Sbjct 274 IGIHG---CIVAGATLALRTKFSASQFWDDCRKYNVTVIQYIGELLRYLCNSPQKPNDRD 330
Query 264 QTQRLGSWSGNPQLRASIGTGLAGALWPVVKKVFNIPRILEFYS 307
RL ++G GL G +W K F I EFY+
Sbjct 331 HKVRL-----------ALGNGLRGDVWRQFVKRFGDICIYEFYA 363
> 7297718
Length=690
Score = 43.1 bits (100), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 44/206 (21%), Positives = 82/206 (39%), Gaps = 31/206 (15%)
Query 156 LYTADLEGHLRATWVSNSR--FLSAGFCWLEAVNLTESDRLLLGVQLSHEVG-LHALAAA 212
+YT+ G +A +S+SR F++AG + + E D + L H G + + +
Sbjct 304 IYTSGTTGLPKAAVISHSRYLFIAAGIHY--TMGFQEEDIFYTPLPLYHTAGGIMCMGQS 361
Query 213 IACKGALLLRSRSSLLSLWPDVKAMDATILWHTGMMWCRLLKAHESYQGGDQTQRLGSWS 272
+ + +R + S + + D +AT+ T + +
Sbjct 362 VLFGSTVSIRKKFSASNYFADCAKYNATV-----------------------TTKPSEYD 398
Query 273 GNPQLRASIGTGLAGALWPVVKKVFNIPRILEFYSPRNLQTPQILFNAWDMLGACAFVPD 332
++R G GL +WP + FNI ++ EFY + + N + +GA FV
Sbjct 399 QKHRVRLVFGNGLRPQIWPQFVQRFNIAKVGEFYGAT--EGNANIMNHDNTVGAIGFVSR 456
Query 333 FAWNSKELERIVEFNEKTGEVYREKN 358
+ I+ + TGE R++N
Sbjct 457 ILPKIYPIS-IIRADPDTGEPIRDRN 481
> Hs13236579
Length=730
Score = 38.5 bits (88), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 48/214 (22%), Positives = 87/214 (40%), Gaps = 26/214 (12%)
Query 153 CCCLYTADLEGHLRATWVSNSRFLSA-GFCWLEAVNLTESDRLLLGVQLSHEVGLHALAA 211
C ++T+ G +A +S+ + L GF L V+ + D + L + L H G +L
Sbjct 330 CLYIFTSGTTGLPKAARISHLKILQCQGFYQLCGVH--QEDVIYLALPLYHMSG--SLLG 385
Query 212 AIACKG---ALLLRSRSSLLSLWPDVKAMDATILWHTGMMWCRLLKAHESYQGGDQTQRL 268
+ C G ++L+S+ S W D + T+ + G + CR L Q + +R
Sbjct 386 IVGCMGIGATVVLKSKFSAGQFWEDCQQHRVTVFQYIGEL-CRYLVN----QPPSKAER- 439
Query 269 GSWSGNPQLRASIGTGLAGALWPVVKKVFNIPRILEFYSPRNLQTPQILFNAWDMLGACA 328
++R ++G+GL W + F ++LE Y I + G
Sbjct 440 -----GHKVRLAVGSGLRPDTWERFVRRFGPLQVLETYGLTEGNVATINYT-----GQRG 489
Query 329 FVPDFAWNSKEL--ERIVEFNEKTGEVYREKNNQ 360
V +W K + ++ ++ TGE R+
Sbjct 490 AVGRASWLYKHIFPFSLIRYDVTTGEPIRDPQGH 523
> Hs13325055
Length=619
Score = 37.4 bits (85), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 35/156 (22%), Positives = 65/156 (41%), Gaps = 13/156 (8%)
Query 153 CCCLYTADLEGHLRATWVSNSRFLSAGFCWLEAVNLTESDRLLLGVQLSHE-VGLHALAA 211
C ++T+ G +A +S + L G L A T D + + + L H + ++
Sbjct 219 CLYIFTSGTTGLPKAAVISQLQVLR-GSAVLWAFGCTAHDIVYITLPLYHSSAAILGISG 277
Query 212 AIACKGALLLRSRSSLLSLWPDVKAMDATILWHTGMMWCRLLKAHESYQGGDQTQRLGSW 271
+ +L+ + S W D K D T+ + G + CR L Q++R G
Sbjct 278 CVELGATCVLKKKFSASQFWSDCKKYDVTVFQYIGEL-CRYL--------CKQSKREG-- 326
Query 272 SGNPQLRASIGTGLAGALWPVVKKVFNIPRILEFYS 307
+ ++R +IG G+ +W F ++ E Y+
Sbjct 327 EKDHKVRLAIGNGIRSDVWREFLDRFGNIKVCELYA 362
> YBR041w
Length=623
Score = 35.8 bits (81), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 40/178 (22%), Positives = 69/178 (38%), Gaps = 13/178 (7%)
Query 152 PCCCLYTADLEGHLRATWVSNSRFLSAGFCWLEAVNLTESDRLLLGVQLSHEVG-LHALA 210
P +YT+ G ++ +S + + +++T + + L H L
Sbjct 251 PSMLIYTSGTTGLPKSAIMSWRKSSVGCQVFGHVLHMTNESTVFTAMPLFHSTAALLGAC 310
Query 211 AAIACKGALLLRSRSSLLSLWPDVKAMDATILWHTGMMWCRLLKAHESYQGGDQTQRLGS 270
A ++ G L L + S + W V AT + + G + CR L H +
Sbjct 311 AILSHGGCLALSHKFSASTFWKQVYLTGATHIQYVGEV-CRYL-LHTP---------ISK 359
Query 271 WSGNPQLRASIGTGLAGALWPVVKKVFNIPRILEFYSPRNLQTPQILFNAWDM-LGAC 327
+ +++ + G GL +W +K FNI I EFY+ F D +GAC
Sbjct 360 YEKMHKVKVAYGNGLRPDIWQDFRKRFNIEVIGEFYAATEAPFATTTFQKGDFGIGAC 417
> CE09790
Length=1153
Score = 32.3 bits (72), Expect = 1.6, Method: Composition-based stats.
Identities = 19/56 (33%), Positives = 25/56 (44%), Gaps = 2/56 (3%)
Query 50 HEDEGQPGSYPCSVYICGLKDAKEQDGERSLIPHVHSIVKE--FLTEEEAAHHRGN 103
HE G P S YIC L + Q RS + +H ++ FL E + HR N
Sbjct 958 HEMAGAPDSTSVDYYICALNFVEYQSLSRSALTKIHGDLESYIFLHELDGDKHRYN 1013
> YER021w
Length=523
Score = 31.6 bits (70), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 12/18 (66%), Positives = 15/18 (83%), Gaps = 0/18 (0%)
Query 333 FAWNSKELERIVEFNEKT 350
F W+SKELE++VEFN K
Sbjct 145 FLWDSKELEQLVEFNRKV 162
> Hs18599609
Length=270
Score = 30.8 bits (68), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 35/71 (49%), Gaps = 9/71 (12%)
Query 296 VFNIPRILEFYSPRNLQTPQILFNAWDMLGACAFVPDFAWNSKELERIVEFNEKTGEVYR 355
V I + E Y + + P+++ + C V F NS EL ++EFN+K ++
Sbjct 65 VLEIVNLRELYQEKKVYCPRMILRSL-----CPCVLKFDDNSTELATLIEFNKK----FQ 115
Query 356 EKNNQEGQGGQ 366
EK +Q + G+
Sbjct 116 EKTHQLIESGR 126
> SPAC1006.06
Length=1158
Score = 30.0 bits (66), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 36/70 (51%), Gaps = 7/70 (10%)
Query 68 LKDAKEQDGERSLIPHVHSIVKEFLTEEEAAHHRGNTENRPN----DEQHKCKPSGFPML 123
LK + + + IP V +++EFLT + + G TENR + +EQ C P+ L
Sbjct 604 LKYTDKDNPDTENIPRVIEMIREFLT--KLNYETGKTENRLSLLQLNEQLSCSPADRAKL 661
Query 124 -LKEPLRKLV 132
L +P R L+
Sbjct 662 TLFDPSRLLI 671
Lambda K H
0.318 0.133 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9331090076
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40