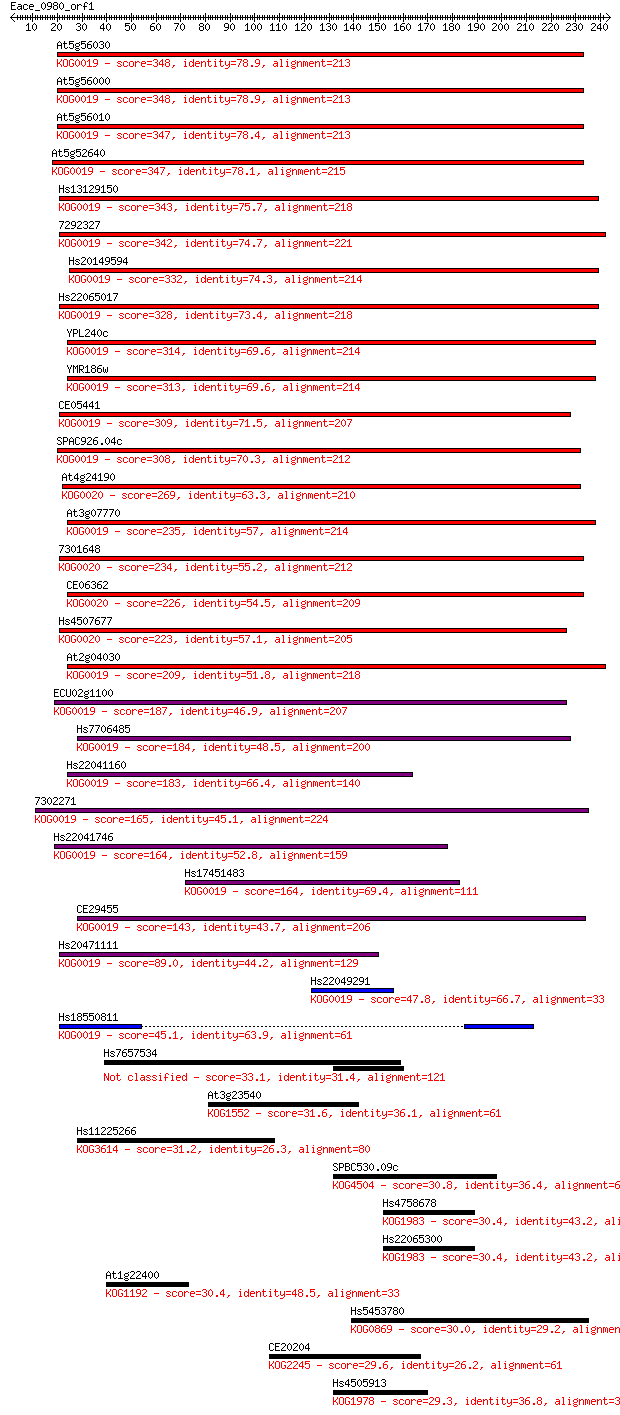
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0980_orf1
Length=243
Score E
Sequences producing significant alignments: (Bits) Value
At5g56030 348 7e-96
At5g56000 348 7e-96
At5g56010 347 1e-95
At5g52640 347 1e-95
Hs13129150 343 2e-94
7292327 342 4e-94
Hs20149594 332 4e-91
Hs22065017 328 7e-90
YPL240c 314 1e-85
YMR186w 313 1e-85
CE05441 309 4e-84
SPAC926.04c 308 7e-84
At4g24190 269 4e-72
At3g07770 235 5e-62
7301648 234 2e-61
CE06362 226 3e-59
Hs4507677 223 4e-58
At2g04030 209 4e-54
ECU02g1100 187 1e-47
Hs7706485 184 2e-46
Hs22041160 183 3e-46
7302271 165 9e-41
Hs22041746 164 1e-40
Hs17451483 164 1e-40
CE29455 143 4e-34
Hs20471111 89.0 8e-18
Hs22049291 47.8 2e-05
Hs18550811 45.1 1e-04
Hs7657534 33.1 0.52
At3g23540 31.6 1.6
Hs11225266 31.2 2.2
SPBC530.09c 30.8 2.7
Hs4758678 30.4 3.7
Hs22065300 30.4 3.7
At1g22400 30.4 3.8
Hs5453780 30.0 4.0
CE20204 29.6 6.5
Hs4505913 29.3 7.5
> At5g56030
Length=699
Score = 348 bits (892), Expect = 7e-96, Method: Compositional matrix adjust.
Identities = 168/213 (78%), Positives = 187/213 (87%), Gaps = 0/213 (0%)
Query 20 MENKETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTK 79
M + ETFAF A+I QL+SLIINTFYSNKEIFLRELISN+SDALDKIR+E++TD KL +
Sbjct 1 MADAETFAFQAEINQLLSLIINTFYSNKEIFLRELISNSSDALDKIRFESLTDKSKLDGQ 60
Query 80 PELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDISMIGQ 139
PELFI +IPDK NNTLTI DSGIGMTKA+LVNNLGTIARSGTK FMEAL AG D+SMIGQ
Sbjct 61 PELFIHIIPDKTNNTLTIIDSGIGMTKADLVNNLGTIARSGTKEFMEALAAGADVSMIGQ 120
Query 140 FGVGFYSAYLVADSVTVVSKHNDDEQYVWESAAGGSFTVQKDDKYEPLGRGTRIILHLKE 199
FGVGFYSAYLVAD V V +KHNDDEQYVWES AGGSFTV +D E LGRGT+++L+LKE
Sbjct 121 FGVGFYSAYLVADKVVVTTKHNDDEQYVWESQAGGSFTVTRDTSGETLGRGTKMVLYLKE 180
Query 200 DQGEYLEERRLKDLVKKHSEFISFPIELAVEKT 232
DQ EYLEERRLKDLVKKHSEFIS+PI L +EKT
Sbjct 181 DQLEYLEERRLKDLVKKHSEFISYPISLWIEKT 213
> At5g56000
Length=699
Score = 348 bits (892), Expect = 7e-96, Method: Compositional matrix adjust.
Identities = 168/213 (78%), Positives = 187/213 (87%), Gaps = 0/213 (0%)
Query 20 MENKETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTK 79
M + ETFAF A+I QL+SLIINTFYSNKEIFLRELISN+SDALDKIR+E++TD KL +
Sbjct 1 MADAETFAFQAEINQLLSLIINTFYSNKEIFLRELISNSSDALDKIRFESLTDKSKLDGQ 60
Query 80 PELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDISMIGQ 139
PELFI +IPDK NNTLTI DSGIGMTKA+LVNNLGTIARSGTK FMEAL AG D+SMIGQ
Sbjct 61 PELFIHIIPDKTNNTLTIIDSGIGMTKADLVNNLGTIARSGTKEFMEALAAGADVSMIGQ 120
Query 140 FGVGFYSAYLVADSVTVVSKHNDDEQYVWESAAGGSFTVQKDDKYEPLGRGTRIILHLKE 199
FGVGFYSAYLVAD V V +KHNDDEQYVWES AGGSFTV +D E LGRGT++IL+LKE
Sbjct 121 FGVGFYSAYLVADKVVVTTKHNDDEQYVWESQAGGSFTVTRDTSGEALGRGTKMILYLKE 180
Query 200 DQGEYLEERRLKDLVKKHSEFISFPIELAVEKT 232
DQ EY+EERRLKDLVKKHSEFIS+PI L +EKT
Sbjct 181 DQMEYIEERRLKDLVKKHSEFISYPISLWIEKT 213
> At5g56010
Length=699
Score = 347 bits (891), Expect = 1e-95, Method: Compositional matrix adjust.
Identities = 167/213 (78%), Positives = 187/213 (87%), Gaps = 0/213 (0%)
Query 20 MENKETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTK 79
M + ETFAF A+I QL+SLIINTFYSNKEIFLRELISN+SDALDKIR+E++TD KL +
Sbjct 1 MADAETFAFQAEINQLLSLIINTFYSNKEIFLRELISNSSDALDKIRFESLTDKSKLDGQ 60
Query 80 PELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDISMIGQ 139
PELFI +IPDK NNTLTI DSGIGMTKA+LVNNLGTIARSGTK FMEAL AG D+SMIGQ
Sbjct 61 PELFIHIIPDKTNNTLTIIDSGIGMTKADLVNNLGTIARSGTKEFMEALAAGADVSMIGQ 120
Query 140 FGVGFYSAYLVADSVTVVSKHNDDEQYVWESAAGGSFTVQKDDKYEPLGRGTRIILHLKE 199
FGVGFYSAYLVAD V V +KHNDDEQYVWES AGGSFTV +D E LGRGT+++L+LKE
Sbjct 121 FGVGFYSAYLVADKVVVTTKHNDDEQYVWESQAGGSFTVTRDTSGEALGRGTKMVLYLKE 180
Query 200 DQGEYLEERRLKDLVKKHSEFISFPIELAVEKT 232
DQ EY+EERRLKDLVKKHSEFIS+PI L +EKT
Sbjct 181 DQMEYIEERRLKDLVKKHSEFISYPISLWIEKT 213
> At5g52640
Length=705
Score = 347 bits (889), Expect = 1e-95, Method: Compositional matrix adjust.
Identities = 168/215 (78%), Positives = 188/215 (87%), Gaps = 0/215 (0%)
Query 18 TKMENKETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLK 77
+M + ETFAF A+I QL+SLIINTFYSNKEIFLRELISN+SDALDKIR+E++TD KL
Sbjct 4 VQMADAETFAFQAEINQLLSLIINTFYSNKEIFLRELISNSSDALDKIRFESLTDKSKLD 63
Query 78 TKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDISMI 137
+PELFIRL+PDK+N TL+I DSGIGMTKA+LVNNLGTIARSGTK FMEALQAG D+SMI
Sbjct 64 GQPELFIRLVPDKSNKTLSIIDSGIGMTKADLVNNLGTIARSGTKEFMEALQAGADVSMI 123
Query 138 GQFGVGFYSAYLVADSVTVVSKHNDDEQYVWESAAGGSFTVQKDDKYEPLGRGTRIILHL 197
GQFGVGFYSAYLVA+ V V +KHNDDEQYVWES AGGSFTV +D EPLGRGT+I L L
Sbjct 124 GQFGVGFYSAYLVAEKVVVTTKHNDDEQYVWESQAGGSFTVTRDVDGEPLGRGTKITLFL 183
Query 198 KEDQGEYLEERRLKDLVKKHSEFISFPIELAVEKT 232
K+DQ EYLEERRLKDLVKKHSEFIS+PI L EKT
Sbjct 184 KDDQLEYLEERRLKDLVKKHSEFISYPIYLWTEKT 218
> Hs13129150
Length=732
Score = 343 bits (879), Expect = 2e-94, Method: Compositional matrix adjust.
Identities = 165/218 (75%), Positives = 192/218 (88%), Gaps = 1/218 (0%)
Query 21 ENKETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTKP 80
E ETFAF A+I QLMSLIINTFYSNKEIFLRELISN+SDALDKIRYE++TDP KL +
Sbjct 15 EEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYESLTDPSKLDSGK 74
Query 81 ELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDISMIGQF 140
EL I LIP+K + TLTI D+GIGMTKA+L+NNLGTIA+SGTKAFMEALQAG DISMIGQF
Sbjct 75 ELHINLIPNKQDRTLTIVDTGIGMTKADLINNLGTIAKSGTKAFMEALQAGADISMIGQF 134
Query 141 GVGFYSAYLVADSVTVVSKHNDDEQYVWESAAGGSFTVQKDDKYEPLGRGTRIILHLKED 200
GVGFYSAYLVA+ VTV++KHNDDEQY WES+AGGSFTV+ D EP+GRGT++ILHLKED
Sbjct 135 GVGFYSAYLVAEKVTVITKHNDDEQYAWESSAGGSFTVRTDTG-EPMGRGTKVILHLKED 193
Query 201 QGEYLEERRLKDLVKKHSEFISFPIELAVEKTHEREVT 238
Q EYLEERR+K++VKKHS+FI +PI L VEK ++EV+
Sbjct 194 QTEYLEERRIKEIVKKHSQFIGYPITLFVEKERDKEVS 231
> 7292327
Length=717
Score = 342 bits (877), Expect = 4e-94, Method: Compositional matrix adjust.
Identities = 165/221 (74%), Positives = 195/221 (88%), Gaps = 1/221 (0%)
Query 21 ENKETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTKP 80
E ETFAF A+I QLMSLIINTFYSNKEIFLRELISNASDALDKIRYE++TDP KL +
Sbjct 3 EEAETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNASDALDKIRYESLTDPSKLDSGK 62
Query 81 ELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDISMIGQF 140
EL+I+LIP+K TLTI D+GIGMTK++LVNNLGTIA+SGTKAFMEALQAG DISMIGQF
Sbjct 63 ELYIKLIPNKTAGTLTIIDTGIGMTKSDLVNNLGTIAKSGTKAFMEALQAGADISMIGQF 122
Query 141 GVGFYSAYLVADSVTVVSKHNDDEQYVWESAAGGSFTVQKDDKYEPLGRGTRIILHLKED 200
GVGFYSAYLVAD VTV SK+NDDEQYVWES+AGGSFTV+ D+ EPLGRGT+I+L++KED
Sbjct 123 GVGFYSAYLVADKVTVTSKNNDDEQYVWESSAGGSFTVRADNS-EPLGRGTKIVLYIKED 181
Query 201 QGEYLEERRLKDLVKKHSEFISFPIELAVEKTHEREVTESE 241
Q +YLEE ++K++V KHS+FI +PI+L VEK E+EV++ E
Sbjct 182 QTDYLEESKIKEIVNKHSQFIGYPIKLLVEKEREKEVSDDE 222
> Hs20149594
Length=724
Score = 332 bits (852), Expect = 4e-91, Method: Compositional matrix adjust.
Identities = 159/214 (74%), Positives = 187/214 (87%), Gaps = 1/214 (0%)
Query 25 TFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTKPELFI 84
TFAF A+I QLMSLIINTFYSNKEIFLRELISNASDALDKIRYE++TDP KL + EL I
Sbjct 14 TFAFQAEIAQLMSLIINTFYSNKEIFLRELISNASDALDKIRYESLTDPSKLDSGKELKI 73
Query 85 RLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDISMIGQFGVGF 144
+IP+ TLT+ D+GIGMTKA+L+NNLGTIA+SGTKAFMEALQAG DISMIGQFGVGF
Sbjct 74 DIIPNPQERTLTLVDTGIGMTKADLINNLGTIAKSGTKAFMEALQAGADISMIGQFGVGF 133
Query 145 YSAYLVADSVTVVSKHNDDEQYVWESAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEY 204
YSAYLVA+ V V++KHNDDEQY WES+AGGSFTV+ D EP+GRGT++ILHLKEDQ EY
Sbjct 134 YSAYLVAEKVVVITKHNDDEQYAWESSAGGSFTVRADHG-EPIGRGTKVILHLKEDQTEY 192
Query 205 LEERRLKDLVKKHSEFISFPIELAVEKTHEREVT 238
LEERR+K++VKKHS+FI +PI L +EK E+E++
Sbjct 193 LEERRVKEVVKKHSQFIGYPITLYLEKEREKEIS 226
> Hs22065017
Length=343
Score = 328 bits (841), Expect = 7e-90, Method: Compositional matrix adjust.
Identities = 160/218 (73%), Positives = 188/218 (86%), Gaps = 1/218 (0%)
Query 21 ENKETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTKP 80
E ETFAF A+I QLMSLIINTFYSNKEIFLRELISN+SDALDKI YE++TDP KL +
Sbjct 15 EEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIWYESLTDPSKLDSGK 74
Query 81 ELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDISMIGQF 140
EL I LIP+K + TLTI D+GIGMTKA+L+NNLGTIA+SGTKAFMEALQAG DISMIGQF
Sbjct 75 ELHINLIPNKQDQTLTIVDTGIGMTKADLINNLGTIAKSGTKAFMEALQAGADISMIGQF 134
Query 141 GVGFYSAYLVADSVTVVSKHNDDEQYVWESAAGGSFTVQKDDKYEPLGRGTRIILHLKED 200
GV FYSAYLVA+ VTV++KHNDDEQY WES+AGGSFTV + D E +GRGT++ILHLKED
Sbjct 135 GVSFYSAYLVAEKVTVITKHNDDEQYAWESSAGGSFTV-RTDTGERMGRGTKVILHLKED 193
Query 201 QGEYLEERRLKDLVKKHSEFISFPIELAVEKTHEREVT 238
Q EYLEE+R+K++VKKHS+ I +PI L VEK ++EV+
Sbjct 194 QTEYLEEQRIKEIVKKHSQLIGYPITLFVEKECDKEVS 231
> YPL240c
Length=709
Score = 314 bits (804), Expect = 1e-85, Method: Compositional matrix adjust.
Identities = 149/214 (69%), Positives = 181/214 (84%), Gaps = 0/214 (0%)
Query 24 ETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTKPELF 83
ETF F A+I QLMSLIINT YSNKEIFLRELISNASDALDKIRY++++DP++L+T+P+LF
Sbjct 4 ETFEFQAEITQLMSLIINTVYSNKEIFLRELISNASDALDKIRYKSLSDPKQLETEPDLF 63
Query 84 IRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDISMIGQFGVG 143
IR+ P L I DSGIGMTKAEL+NNLGTIA+SGTKAFMEAL AG D+SMIGQFGVG
Sbjct 64 IRITPKPEQKVLEIRDSGIGMTKAELINNLGTIAKSGTKAFMEALSAGADVSMIGQFGVG 123
Query 144 FYSAYLVADSVTVVSKHNDDEQYVWESAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGE 203
FYS +LVAD V V+SK NDDEQY+WES AGGSFTV D+ E +GRGT + L LK+DQ E
Sbjct 124 FYSLFLVADRVQVISKSNDDEQYIWESNAGGSFTVTLDEVNERIGRGTILRLFLKDDQLE 183
Query 204 YLEERRLKDLVKKHSEFISFPIELAVEKTHEREV 237
YLEE+R+K+++K+HSEF+++PI+L V K E+EV
Sbjct 184 YLEEKRIKEVIKRHSEFVAYPIQLVVTKEVEKEV 217
> YMR186w
Length=705
Score = 313 bits (803), Expect = 1e-85, Method: Compositional matrix adjust.
Identities = 149/214 (69%), Positives = 182/214 (85%), Gaps = 0/214 (0%)
Query 24 ETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTKPELF 83
ETF F A+I QLMSLIINT YSNKEIFLRELISNASDALDKIRY+A++DP++L+T+P+LF
Sbjct 4 ETFEFQAEITQLMSLIINTVYSNKEIFLRELISNASDALDKIRYQALSDPKQLETEPDLF 63
Query 84 IRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDISMIGQFGVG 143
IR+ P L I DSGIGMTKAEL+NNLGTIA+SGTKAFMEAL AG D+SMIGQFGVG
Sbjct 64 IRITPKPEEKVLEIRDSGIGMTKAELINNLGTIAKSGTKAFMEALSAGADVSMIGQFGVG 123
Query 144 FYSAYLVADSVTVVSKHNDDEQYVWESAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGE 203
FYS +LVAD V V+SK+N+DEQY+WES AGGSFTV D+ E +GRGT + L LK+DQ E
Sbjct 124 FYSLFLVADRVQVISKNNEDEQYIWESNAGGSFTVTLDEVNERIGRGTVLRLFLKDDQLE 183
Query 204 YLEERRLKDLVKKHSEFISFPIELAVEKTHEREV 237
YLEE+R+K+++K+HSEF+++PI+L V K E+EV
Sbjct 184 YLEEKRIKEVIKRHSEFVAYPIQLLVTKEVEKEV 217
> CE05441
Length=702
Score = 309 bits (791), Expect = 4e-84, Method: Compositional matrix adjust.
Identities = 148/207 (71%), Positives = 180/207 (86%), Gaps = 1/207 (0%)
Query 21 ENKETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTKP 80
EN ETFAF A+I QLMSLIINTFYSNKEI+LRELISNASDALDKIRY+A+T+P +L T
Sbjct 3 ENAETFAFQAEIAQLMSLIINTFYSNKEIYLRELISNASDALDKIRYQALTEPSELDTGK 62
Query 81 ELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDISMIGQF 140
ELFI++ P+K TLTI D+GIGMTKA+LVNNLGTIA+SGTKAFMEALQAG DISMIGQF
Sbjct 63 ELFIKITPNKEEKTLTIMDTGIGMTKADLVNNLGTIAKSGTKAFMEALQAGADISMIGQF 122
Query 141 GVGFYSAYLVADSVTVVSKHNDDEQYVWESAAGGSFTVQKDDKYEPLGRGTRIILHLKED 200
GVGFYSA+LVAD V V SK+NDD+ Y WES+AGGSF V+ + E + RGT+I++H+KED
Sbjct 123 GVGFYSAFLVADKVVVTSKNNDDDSYQWESSAGGSFVVRPFNDPE-VTRGTKIVMHIKED 181
Query 201 QGEYLEERRLKDLVKKHSEFISFPIEL 227
Q ++LEER++K++VKKHS+FI +PI+L
Sbjct 182 QIDFLEERKIKEIVKKHSQFIGYPIKL 208
> SPAC926.04c
Length=704
Score = 308 bits (789), Expect = 7e-84, Method: Compositional matrix adjust.
Identities = 149/212 (70%), Positives = 172/212 (81%), Gaps = 0/212 (0%)
Query 20 MENKETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTK 79
M N ETF F A+I QLMSLIINT YSNKEIFLRELISNASDALDKIRY++++DP L +
Sbjct 1 MSNTETFKFEAEISQLMSLIINTVYSNKEIFLRELISNASDALDKIRYQSLSDPHALDAE 60
Query 80 PELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDISMIGQ 139
+LFIR+ PDK N L+I D+GIGMTK +L+NNLG IA+SGTK FMEA +G DISMIGQ
Sbjct 61 KDLFIRITPDKENKILSIRDTGIGMTKNDLINNLGVIAKSGTKQFMEAAASGADISMIGQ 120
Query 140 FGVGFYSAYLVADSVTVVSKHNDDEQYVWESAAGGSFTVQKDDKYEPLGRGTRIILHLKE 199
FGVGFYSAYLVAD V VVSKHNDDEQY+WES+AGGSFTV D L RGT I L +KE
Sbjct 121 FGVGFYSAYLVADKVQVVSKHNDDEQYIWESSAGGSFTVTLDTDGPRLLRGTEIRLFMKE 180
Query 200 DQGEYLEERRLKDLVKKHSEFISFPIELAVEK 231
DQ +YLEE+ +KD VKKHSEFIS+PI+L V +
Sbjct 181 DQLQYLEEKTIKDTVKKHSEFISYPIQLVVTR 212
> At4g24190
Length=823
Score = 269 bits (687), Expect = 4e-72, Method: Compositional matrix adjust.
Identities = 133/214 (62%), Positives = 165/214 (77%), Gaps = 6/214 (2%)
Query 22 NKETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKL----K 77
N E F F A++ +LM +IIN+ YSNK+IFLRELISNASDALDKIR+ A+TD + L
Sbjct 75 NAEKFEFQAEVSRLMDIIINSLYSNKDIFLRELISNASDALDKIRFLALTDKDVLGEGDT 134
Query 78 TKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDISMI 137
K E+ I+L DKA L+I D GIGMTK +L+ NLGTIA+SGT AF+E +Q+ GD+++I
Sbjct 135 AKLEIQIKL--DKAKKILSIRDRGIGMTKEDLIKNLGTIAKSGTSAFVEKMQSSGDLNLI 192
Query 138 GQFGVGFYSAYLVADSVTVVSKHNDDEQYVWESAAGGSFTVQKDDKYEPLGRGTRIILHL 197
GQFGVGFYSAYLVAD + V+SKHNDD QYVWES A G F V +D EPLGRGT I LHL
Sbjct 193 GQFGVGFYSAYLVADYIEVISKHNDDSQYVWESKANGKFAVSEDTWNEPLGRGTEIRLHL 252
Query 198 KEDQGEYLEERRLKDLVKKHSEFISFPIELAVEK 231
+++ GEYLEE +LK+LVK++SEFI+FPI L K
Sbjct 253 RDEAGEYLEESKLKELVKRYSEFINFPISLWASK 286
> At3g07770
Length=803
Score = 235 bits (600), Expect = 5e-62, Method: Compositional matrix adjust.
Identities = 122/221 (55%), Positives = 161/221 (72%), Gaps = 7/221 (3%)
Query 24 ETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTKPELF 83
E F + A++ +LM LI+N+ YSNKE+FLRELISNASDALDK+RY ++T+PE K P+L
Sbjct 95 EKFEYQAEVSRLMDLIVNSLYSNKEVFLRELISNASDALDKLRYLSVTNPELSKDAPDLD 154
Query 84 IRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQ----AGGDISMIGQ 139
IR+ DK N +T+ DSGIGMT+ ELV+ LGTIA+SGT FM+AL+ AGGD ++IGQ
Sbjct 155 IRIYADKENGIITLTDSGIGMTRQELVDCLGTIAQSGTAKFMKALKDSKDAGGDNNLIGQ 214
Query 140 FGVGFYSAYLVADSVTVVSKH-NDDEQYVWESAA-GGSFTVQKD-DKYEPLGRGTRIILH 196
FGVGFYSA+LVAD V V +K D+QYVWE A SFT+Q+D D + RGTRI LH
Sbjct 215 FGVGFYSAFLVADRVIVSTKSPKSDKQYVWEGEANSSSFTIQEDTDPQSLIPRGTRITLH 274
Query 197 LKEDQGEYLEERRLKDLVKKHSEFISFPIELAVEKTHEREV 237
LK++ + + R++ LVK +S+F+SFPI EK + +EV
Sbjct 275 LKQEAKNFADPERIQKLVKNYSQFVSFPIYTWQEKGYTKEV 315
> 7301648
Length=787
Score = 234 bits (596), Expect = 2e-61, Method: Compositional matrix adjust.
Identities = 117/218 (53%), Positives = 161/218 (73%), Gaps = 7/218 (3%)
Query 21 ENKETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTKP 80
E + F F ++ ++M LIIN+ Y NKEIFLRELISNASDA+DKIR A+++ ++L+T P
Sbjct 70 EKAKKFTFQTEVNRMMKLIINSLYRNKEIFLRELISNASDAIDKIRLLALSNSKELETNP 129
Query 81 ELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQ-----AGGDIS 135
EL IR+ DK N L I DSGIGMT +L+NNLGTIA+SGT F+ +Q G D++
Sbjct 130 ELHIRIKADKENKALHIMDSGIGMTHQDLINNLGTIAKSGTADFLAKMQDPSKSEGLDMN 189
Query 136 -MIGQFGVGFYSAYLVADSVTVVSKHNDDEQYVWESAAGGSFTVQKDDKYEPLGRGTRII 194
MIGQFGVGFYSA+LVAD V V +KHNDD+QY+WES A SF++ +D + + L RG+ I
Sbjct 190 DMIGQFGVGFYSAFLVADRVVVTTKHNDDKQYIWESDA-NSFSITEDPRGDTLKRGSVIS 248
Query 195 LHLKEDQGEYLEERRLKDLVKKHSEFISFPIELAVEKT 232
L+LKE+ ++LEE +++L++K+S+FI+FPI + KT
Sbjct 249 LYLKEEAQDFLEEDTVRELIRKYSQFINFPIRMWSSKT 286
> CE06362
Length=760
Score = 226 bits (576), Expect = 3e-59, Method: Compositional matrix adjust.
Identities = 114/214 (53%), Positives = 153/214 (71%), Gaps = 6/214 (2%)
Query 24 ETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTKPELF 83
E F A++ ++M LIIN+ Y NKEIFLRELISNASDALDKIR ++TDPE+L+ E+
Sbjct 62 EKHEFQAEVNRMMKLIINSLYRNKEIFLRELISNASDALDKIRLLSLTDPEQLRETEEMS 121
Query 84 IRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDIS-----MIG 138
+++ D+ N L I D+G+GMT+ +L+NNLGTIARSGT F+ L S +IG
Sbjct 122 VKIKADRENRLLHITDTGVGMTRQDLINNLGTIARSGTSEFLSKLMDTATSSDQQQDLIG 181
Query 139 QFGVGFYSAYLVADSVTVVSKHNDDEQYVWESAAGGSFTVQKDDKYEPLGRGTRIILHLK 198
QFGVGFY+A+LVAD V V +K+NDD+QY+WES + SFT+ KD + L RGT+I L+LK
Sbjct 182 QFGVGFYAAFLVADRVVVTTKNNDDDQYIWESDS-ASFTISKDPRGNTLKRGTQITLYLK 240
Query 199 EDQGEYLEERRLKDLVKKHSEFISFPIELAVEKT 232
E+ ++LE LK+LV K+S+FI+F I L KT
Sbjct 241 EEAADFLEPDTLKNLVHKYSQFINFDIFLWQSKT 274
> Hs4507677
Length=803
Score = 223 bits (567), Expect = 4e-58, Method: Compositional matrix adjust.
Identities = 117/210 (55%), Positives = 149/210 (70%), Gaps = 6/210 (2%)
Query 21 ENKETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTKP 80
E E FAF A++ ++M LIIN+ Y NKEIFLRELISNASDALDKIR ++TD L
Sbjct 71 EKSEKFAFQAEVNRMMKLIINSLYKNKEIFLRELISNASDALDKIRLISLTDENALSGNE 130
Query 81 ELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFM----EALQAGGDIS- 135
EL +++ DK N L + D+G+GMT+ ELV NLGTIA+SGT F+ EA + G S
Sbjct 131 ELTVKIKCDKEKNLLHVTDTGVGMTREELVKNLGTIAKSGTSEFLNKMTEAQEDGQSTSE 190
Query 136 MIGQFGVGFYSAYLVADSVTVVSKHNDDEQYVWESAAGGSFTVQKDDKYEPLGRGTRIIL 195
+IGQFGVGFYSA+LVAD V V SKHN+D Q++WES + F+V D + LGRGT I L
Sbjct 191 LIGQFGVGFYSAFLVADKVIVTSKHNNDTQHIWESDS-NEFSVIADPRGNTLGRGTTITL 249
Query 196 HLKEDQGEYLEERRLKDLVKKHSEFISFPI 225
LKE+ +YLE +K+LVKK+S+FI+FPI
Sbjct 250 VLKEEASDYLELDTIKNLVKKYSQFINFPI 279
> At2g04030
Length=780
Score = 209 bits (532), Expect = 4e-54, Method: Compositional matrix adjust.
Identities = 113/226 (50%), Positives = 156/226 (69%), Gaps = 8/226 (3%)
Query 24 ETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTKPELF 83
E F + A++ +L+ LI+++ YS+KE+FLREL+SNASDALDK+R+ ++T+P L +L
Sbjct 77 EKFEYQAEVSRLLDLIVHSLYSHKEVFLRELVSNASDALDKLRFLSVTEPSLLGDGGDLE 136
Query 84 IRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQA----GGDISMIGQ 139
IR+ PD N T+TI D+GIGMTK EL++ LGTIA+SGT F++AL+ G D +IGQ
Sbjct 137 IRIKPDPDNGTITITDTGIGMTKEELIDCLGTIAQSGTSKFLKALKENKDLGADNGLIGQ 196
Query 140 FGVGFYSAYLVADSVTVVSKH-NDDEQYVWESAAGGS--FTVQKDDKYEPLGRGTRIILH 196
FGVGFYSA+LVA+ V V +K D+QYVWES A S ++ D L RGT+I L+
Sbjct 197 FGVGFYSAFLVAEKVVVSTKSPKSDKQYVWESVADSSSYLIREETDPDNILRRGTQITLY 256
Query 197 LKEDQG-EYLEERRLKDLVKKHSEFISFPIELAVEKTHEREVTESE 241
L+ED E+ E R+K+LVK +S+F+ FPI EK+ EV E E
Sbjct 257 LREDDKYEFAESTRIKNLVKNYSQFVGFPIYTWQEKSRTIEVEEDE 302
> ECU02g1100
Length=690
Score = 187 bits (476), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 97/214 (45%), Positives = 145/214 (67%), Gaps = 8/214 (3%)
Query 19 KMENKETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEK--- 75
K ++ ET F D+ Q+M +I + YS+KE+FLREL+SN+SDA DK++ EK
Sbjct 14 KDKHSETHGFEVDVNQMMDTMIKSVYSSKELFLRELVSNSSDACDKLKALYFQLREKGCV 73
Query 76 LKTKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDIS 135
L L I +IP+K N TLTI+D+GIGMTK +L+N +GTIA SGTK F E ++ G+ +
Sbjct 74 LDPVTSLGIEIIPNKDNRTLTIKDNGIGMTKPDLMNFIGTIASSGTKKFREEMKEKGNSA 133
Query 136 ----MIGQFGVGFYSAYLVADSVTVVSKHNDDEQYVWESAAGGSFTVQKDDKYEPLGRGT 191
+IGQFG+GFYS+YLVA+ V +++KH DE VW S +T+++ D EP GT
Sbjct 134 DASNLIGQFGLGFYSSYLVAERVDLITKHPSDEALVWTSTGRDVYTIEEYDG-EPFAHGT 192
Query 192 RIILHLKEDQGEYLEERRLKDLVKKHSEFISFPI 225
++L++KE + E+L+ +R+ ++VKK+S F+ +PI
Sbjct 193 SLVLYIKEGEEEFLDPKRISEIVKKYSLFVFYPI 226
> Hs7706485
Length=704
Score = 184 bits (466), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 97/204 (47%), Positives = 139/204 (68%), Gaps = 10/204 (4%)
Query 28 FNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTKPELFIRLI 87
F A+ ++L+ ++ + YS KE+F+RELISNASDAL+K+R++ ++D + L PE+ I L
Sbjct 90 FQAETKKLLDIVARSLYSEKEVFIRELISNASDALEKLRHKLVSDGQAL---PEMEIHLQ 146
Query 88 PDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDIS--MIGQFGVGFY 145
+ T+TI+D+GIGMT+ ELV+NLGTIARSG+KAF++ALQ + S +IGQFGVGFY
Sbjct 147 TNAEKGTITIQDTGIGMTQEELVSNLGTIARSGSKAFLDALQNQAEASSKIIGQFGVGFY 206
Query 146 SAYLVADSVTVVSKHNDDEQ--YVWESAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGE 203
SA++VAD V V S+ Y W S G F + + GT+II+HLK D E
Sbjct 207 SAFMVADRVEVYSRSAAPGSLGYQWLSDGSGVFEIAEASGVRT---GTKIIIHLKSDCKE 263
Query 204 YLEERRLKDLVKKHSEFISFPIEL 227
+ E R++D+V K+S F+SFP+ L
Sbjct 264 FSSEARVRDVVTKYSNFVSFPLYL 287
> Hs22041160
Length=343
Score = 183 bits (464), Expect = 3e-46, Method: Compositional matrix adjust.
Identities = 93/140 (66%), Positives = 109/140 (77%), Gaps = 13/140 (9%)
Query 24 ETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTKPELF 83
ETFAF A+I QLMSLIINTFYSNKEIFL ELISNASDALDKIRYE++TDP KL + EL
Sbjct 13 ETFAFQAEIAQLMSLIINTFYSNKEIFLWELISNASDALDKIRYESLTDPSKLDSGKELK 72
Query 84 IRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDISMIGQFGVG 143
I +IP+ +TLT+ D+GIGMTKA+L+NNLGTIA+ ISMIGQFG+G
Sbjct 73 IDIIPNTQEHTLTLVDTGIGMTKADLINNLGTIAKF-------------HISMIGQFGIG 119
Query 144 FYSAYLVADSVTVVSKHNDD 163
FYSAYLVA+ V V++K NDD
Sbjct 120 FYSAYLVAEKVVVITKPNDD 139
> 7302271
Length=691
Score = 165 bits (417), Expect = 9e-41, Method: Compositional matrix adjust.
Identities = 101/243 (41%), Positives = 148/243 (60%), Gaps = 23/243 (9%)
Query 11 GTLYFRVTKMENKETFA-------FNADIQQLMSLIINTFYSNKEIFLRELISNASDALD 63
G L R E K+ F A+ +QL+ ++ + YS+ E+F+RELISNASDAL+
Sbjct 44 GALSLRRYSTETKQASGSVVDKHEFQAETRQLLDIVARSLYSDHEVFVRELISNASDALE 103
Query 64 KIRYEAIT-DPEKL--KTKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSG 120
K RY +++ E L K +P L IR+ DK L I+D+GIGMTK ELV+NLGTIARSG
Sbjct 104 KFRYTSLSAGGENLAGKDRP-LEIRITTDKPLMQLIIQDTGIGMTKEELVSNLGTIARSG 162
Query 121 TKAFMEAL---QAG----GDISMIGQFGVGFYSAYLVADSVTVVSKHN--DDEQYVWESA 171
+K F+E + Q G ++IGQFGVGFYS+++VA+ V V ++ + W +
Sbjct 163 SKKFLEQMKGTQQGASSEASSNIIGQFGVGFYSSFIVANKVEVFTRAAVPNAPGLRWSTD 222
Query 172 AGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELAVEK 231
G++ +++ E GTRI+LHLK D EY +E R+K ++KK+S F+ PI L ++
Sbjct 223 GSGTYEIEEVPDVE---LGTRIVLHLKTDCREYADEERIKAVIKKYSNFVGSPILLNGKQ 279
Query 232 THE 234
+E
Sbjct 280 ANE 282
> Hs22041746
Length=1595
Score = 164 bits (416), Expect = 1e-40, Method: Composition-based stats.
Identities = 84/161 (52%), Positives = 110/161 (68%), Gaps = 2/161 (1%)
Query 19 KMENKETFAFNADIQQLMSLIINTFYSN--KEIFLRELISNASDALDKIRYEAITDPEKL 76
K E ++ +L +L++ ++ + L E + NA+ L + E++TDP KL
Sbjct 601 KHPGAEHTSWGVAFSELCALVVGILLTSCHRHSRLAETMPNANLILPCLLPESLTDPSKL 660
Query 77 KTKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDISM 136
+ E I LIP+K + TLTI D+GIGMTKA+L+NNLGTI +S TK FME LQAG DISM
Sbjct 661 DSGKEPHISLIPNKQDRTLTIVDTGIGMTKADLINNLGTITKSETKVFMEVLQAGADISM 720
Query 137 IGQFGVGFYSAYLVADSVTVVSKHNDDEQYVWESAAGGSFT 177
IGQF VGFYSAY VA+ VTV++KHN+DEQY WES+ GSFT
Sbjct 721 IGQFSVGFYSAYSVAEKVTVITKHNNDEQYAWESSLRGSFT 761
> Hs17451483
Length=112
Score = 164 bits (416), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 77/111 (69%), Positives = 90/111 (81%), Gaps = 0/111 (0%)
Query 72 DPEKLKTKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAG 131
D KL + L I L+P+K N TLTI D+ IGMTK +L+NNLGT+ +SGTKAFMEALQA
Sbjct 2 DSSKLDSGKGLHINLVPNKQNQTLTIVDTRIGMTKTDLINNLGTVTKSGTKAFMEALQAD 61
Query 132 GDISMIGQFGVGFYSAYLVADSVTVVSKHNDDEQYVWESAAGGSFTVQKDD 182
DISMIG FGVGFYSAYLVA VTV+SKHN+DEQY WES+AGGSFTV+ D+
Sbjct 62 ADISMIGHFGVGFYSAYLVAKKVTVISKHNNDEQYTWESSAGGSFTVRTDE 112
> CE29455
Length=657
Score = 143 bits (360), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 90/213 (42%), Positives = 131/213 (61%), Gaps = 17/213 (7%)
Query 28 FNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTKPELFIRLI 87
F A+ + LM ++ + YS+ E+F+RELISNASDAL+K RY + + P IR+
Sbjct 32 FQAETRNLMDIVAKSLYSHSEVFVRELISNASDALEKRRYAELKG--DVAEGPSE-IRIT 88
Query 88 PDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDISMIGQFGVGFYSA 147
+K T+T ED+GIGM + +LV LGTIA+SG+K F+E + + ++IGQFGVGFYSA
Sbjct 89 TNKDKRTITFEDTGIGMNREDLVKFLGTIAKSGSKDFIENNKENAE-AVIGQFGVGFYSA 147
Query 148 YLVADSVTVVSKHNDDEQYVWESAAGG-SFTVQKDDKYE-----PLGRGTRIILHLK-ED 200
++VADSV V ++ V S A G +T D+ YE L GT+I + LK D
Sbjct 148 FMVADSVVVTTRK------VGSSDADGLQWTWNGDNSYEIAETSGLQTGTKIEIRLKVGD 201
Query 201 QGEYLEERRLKDLVKKHSEFISFPIELAVEKTH 233
Y EE R+K+++ K+S F+S PI + E+ +
Sbjct 202 SATYAEEDRIKEVINKYSYFVSAPILVNGERVN 234
> Hs20471111
Length=217
Score = 89.0 bits (219), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 57/129 (44%), Positives = 62/129 (48%), Gaps = 50/129 (38%)
Query 21 ENKETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTKP 80
E E F A I + MSLIINTFY NKEIFL +
Sbjct 6 EEVEICTFQALIAEPMSLIINTFYFNKEIFLWK--------------------------- 38
Query 81 ELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDISMIGQF 140
R + D L+NNLGTIA+SGTKAFMEALQAG ISMIGQF
Sbjct 39 ----RFLAD-------------------LINNLGTIAKSGTKAFMEALQAGARISMIGQF 75
Query 141 GVGFYSAYL 149
G GFY AYL
Sbjct 76 GFGFYCAYL 84
> Hs22049291
Length=131
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 22/33 (66%), Positives = 24/33 (72%), Gaps = 0/33 (0%)
Query 123 AFMEALQAGGDISMIGQFGVGFYSAYLVADSVT 155
F EAL G DISMIG GVGFYSAYL+A+S
Sbjct 98 GFREALPTGADISMIGWVGVGFYSAYLMAESCA 130
> Hs18550811
Length=329
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 19/28 (67%), Positives = 24/28 (85%), Gaps = 0/28 (0%)
Query 185 EPLGRGTRIILHLKEDQGEYLEERRLKD 212
EP R T++ILHLKEDQ EYLEER++K+
Sbjct 43 EPTSRDTKVILHLKEDQIEYLEERQIKE 70
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/33 (60%), Positives = 26/33 (78%), Gaps = 0/33 (0%)
Query 21 ENKETFAFNADIQQLMSLIINTFYSNKEIFLRE 53
E ETFAF A++ Q M L INTFYSN++IFL++
Sbjct 10 EEVETFAFQAEVAQFMYLNINTFYSNQKIFLQD 42
> Hs7657534
Length=3829
Score = 33.1 bits (74), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 38/131 (29%), Positives = 53/131 (40%), Gaps = 36/131 (27%)
Query 39 IINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTKPELFIRLIPDKA------- 91
I+N + S KE+ L+EL+ NA DA + DP + R+ DK
Sbjct 1785 ILNAYPSEKEM-LKELLQNADDA-KATEICFVFDPRQHPVD-----RIFDDKWAPLQGPA 1837
Query 92 ----NNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDISMIGQFGVGFYSA 147
NN ED G + NLG GTK G+ GQ+G+GF S
Sbjct 1838 LCVYNNQPFTEDDVRG------IQNLG----KGTKE--------GNPYKTGQYGIGFNSV 1879
Query 148 YLVADSVTVVS 158
Y + D + +S
Sbjct 1880 YHITDCPSFIS 1890
Score = 30.0 bits (66), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 10/28 (35%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 132 GDISMIGQFGVGFYSAYLVADSVTVVSK 159
G++ +G+FG+GF S Y + D ++S+
Sbjct 798 GEVDKVGKFGLGFNSVYHITDIPIIMSR 825
> At3g23540
Length=568
Score = 31.6 bits (70), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 33/61 (54%), Gaps = 2/61 (3%)
Query 81 ELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTIARSGTKAFMEALQAGGDISMIGQF 140
E I L+P T++ SG G++ E V LG + KA +E L+ G+IS+IG +
Sbjct 80 EAAIVLLPSNIT-VFTLDFSGSGLSGGEHVT-LGWNEKDDLKAVVEFLRQDGNISLIGLW 137
Query 141 G 141
G
Sbjct 138 G 138
> Hs11225266
Length=1165
Score = 31.2 bits (69), Expect = 2.2, Method: Composition-based stats.
Identities = 21/85 (24%), Positives = 41/85 (48%), Gaps = 7/85 (8%)
Query 28 FNADIQ----QLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKTKPELF 83
FN D++ L ++++ SNK F+R + N +D D + Y + + + ++ L
Sbjct 369 FNGDVEWKSCDLEEVMVDALVSNKPEFVRLFVDNGADVADFLTYGRLQELYRSVSRKSLL 428
Query 84 IRLIPDKANNT-LTIEDSGIGMTKA 107
L+ K LT+ +G+G +A
Sbjct 429 FDLLQRKQEEARLTL--AGLGTQQA 451
> SPBC530.09c
Length=296
Score = 30.8 bits (68), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 36/74 (48%), Gaps = 9/74 (12%)
Query 132 GDISMIG-QFGVGFYSAYLVADSVTVVSKHNDDEQYVWESAAGGSFTVQKDD-------K 183
GD S+ G FG F S L V+ ++ + + E + GG FTV+ DD
Sbjct 69 GDYSVNGYDFGTNF-SINLCHPVVSNLTNYTVEGDVSSEDSIGGFFTVEDDDLYSIGQAA 127
Query 184 YEPLGRGTRIILHL 197
Y+P RG ++I+ L
Sbjct 128 YKPYFRGKKLIMQL 141
> Hs4758678
Length=1057
Score = 30.4 bits (67), Expect = 3.7, Method: Composition-based stats.
Identities = 16/37 (43%), Positives = 18/37 (48%), Gaps = 0/37 (0%)
Query 152 DSVTVVSKHNDDEQYVWESAAGGSFTVQKDDKYEPLG 188
DS TVVS H+D VW AG T+Q P G
Sbjct 249 DSSTVVSSHSDGSYAVWSVDAGSFPTLQPTVATTPYG 285
> Hs22065300
Length=1062
Score = 30.4 bits (67), Expect = 3.7, Method: Composition-based stats.
Identities = 16/37 (43%), Positives = 18/37 (48%), Gaps = 0/37 (0%)
Query 152 DSVTVVSKHNDDEQYVWESAAGGSFTVQKDDKYEPLG 188
DS TVVS H+D VW AG T+Q P G
Sbjct 248 DSSTVVSSHSDGSYAVWSVDAGSFPTLQPTVATTPYG 284
> At1g22400
Length=489
Score = 30.4 bits (67), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 16/33 (48%), Positives = 19/33 (57%), Gaps = 1/33 (3%)
Query 40 INTFYSNKEIFLRELISNASDALDKIRYEAITD 72
+NT Y N FLR SNA D L R+E+I D
Sbjct 45 VNTVY-NHNRFLRSRGSNALDGLPSFRFESIAD 76
> Hs5453780
Length=207
Score = 30.0 bits (66), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 28/99 (28%), Positives = 47/99 (47%), Gaps = 8/99 (8%)
Query 139 QFGVGFYSAYLVADSVTVVSKHNDDEQYV--WESAAGGSFTVQKDDKYEPLGRGTRIILH 196
Q G+ SA + S V+ H+D E + E G + ++ D Y P+ RI+ +
Sbjct 14 QLGI---SADYIGGSHYVIQPHDDTEDSMNDHEDTNGSKESFREQDIYLPIANVARIMKN 70
Query 197 LKEDQGEYLEERRLKDLVKKH-SEFISFPIELAVEKTHE 234
G+ ++ K+ V++ SEFISF A E+ H+
Sbjct 71 AIPQTGKIAKD--AKECVQECVSEFISFITSEASERCHQ 107
> CE20204
Length=655
Score = 29.6 bits (65), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 16/68 (23%), Positives = 31/68 (45%), Gaps = 7/68 (10%)
Query 106 KAELVNNLGTIARSGTKAFM-------EALQAGGDISMIGQFGVGFYSAYLVADSVTVVS 158
+ E++ NL + + K E + AGG + G + +G +S+ D++ VV
Sbjct 53 RMEVLRNLNRLVKEWVKNVTAMKIPNGEGVNAGGKLFTFGSYRLGVHSSGADIDTLAVVP 112
Query 159 KHNDDEQY 166
+H D +
Sbjct 113 RHIDRSDF 120
> Hs4505913
Length=862
Score = 29.3 bits (64), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 23/38 (60%), Gaps = 1/38 (2%)
Query 132 GDISMIGQFGVGFYSAYLVADSVTVVSKHNDDEQYVWE 169
++ +IGQF +GF L D + +V +H DE+Y +E
Sbjct 674 AEMEIIGQFNLGFIITKLNED-IFIVDQHATDEKYNFE 710
Lambda K H
0.316 0.134 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4867742342
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40